CYNAPSE: supporting health data research
Large amounts of information are generated, and must be safely stored, during our medical research programmes on and around the Cambridge Biomedical Campus.
CYNAPSE is a project, funded through the NIHR Cambridge BRC, which aims to build a secure infrastructure allowing Cambridge-based researchers to accountably and efficiently use health data to support critical studies.
How will CYNAPSE support important new research?
Medical research often involves the creation of large amounts of genetic and other biological data, potentially offering important insights into health and disease.
Currently, these large, important datasets are stored in many locations, and their format may differ. Variations in the way that data is stored and accessed makes it challenging for different research groups to use each other’s data, even within the same organisation.
The CYNAPSE team will also ensure that access to data is safely and fairly granted through new information governance systems.
You can read more about CYNAPSE and how we have worked with patients and the public to develop it here.
The NIHR Cambridge BRC, working with CYNAPSE, are committed to supporting a new, standardised, approach to securely storing and sharing medical information. This will allow approved scientists to work with each other’s information more easily, increasing the potential benefit to patients of each dataset.
CYNAPSE – frequently asked questions
Introducing CYNAPSE
Many modern health research studies, such as the Genomics England 100,000 Genomes project and the UK Biobank, collect and investigate biological samples from many thousands of people.
This research creates vast amounts of data about their genetics and other biological information and how this relates to health and disease. The very large, very valuable datasets produced by these, and other projects, are currently held in many locations, and each differs in the way that it is recorded and stored. This makes it challenging for different research groups to make use of each other’s data, even within the same organisation.
If data were held and saved in a standardised way, scientists could work with each other’s information more easily, maximising the usefulness – and potential benefit to patients – of each dataset.
CYNAPSE aims to improve this situation locally by developing improved data connection systems (known as ‘infrastructure’) across the Cambridge Biomedical Campus, providing a standardised system for researchers to store and access data so that information can be shared and used more efficiently.
Developing improved oversight (‘governance’) processes is an important part of CYNAPSE too, ensuring that data are well-managed and fairly accessed, including by research groups that don’t have access to large computational resources.
The CYNAPSE team are working with the company Lifebit, who will build this new software platform and cloud-based TRE (Lifebit CloudOS).
How does this fit into the DARE UK Federated Genomics project?
Many research institutions and data providers use computing environments known as ‘trusted research environments’ (TREs) as a way to safely analyse data.
However, these individual TREs currently cannot easily be used in combination. Each organisation will have its own governance policies that its TREs operate within and moving data from one TRE into another is costly and time consuming due to the need to navigate the data governance policies of multiple organisations. The ability to analyse datasets held across different organisations could be very valuable in supporting new research.
Part of the CYNAPSE programme will allow for connectivity between datasets amongst different research organisations without the need to move datasets to new locations (known as ‘federation’). We will do this by developing technology to allow datasets held in these independent locations to be analysed simultaneously. The results of the separate analyses can then be combined without the original data ever having to move.
The initial stage of this part of the programme is being enabled through the Multi-party trusted research environment federation: Establishing infrastructure for secure analysis across different clinical-genomic datasets, one of nine short-term projects funded by UK Research & Innovation as part of Phase 1 of the DARE UK (Data and Analytics Research Environments UK) programme, delivered in partnership with Health Data Research UK (HDR UK) and ADR UK (Administrative Data Research UK).
This proof of concept project will test whether researchers can access and simultaneously analyse data that is held in the two protected areas of CYNAPSE and Genomics England.
The figure below shows where the DARE UK sprint project fits into the CYNAPSE 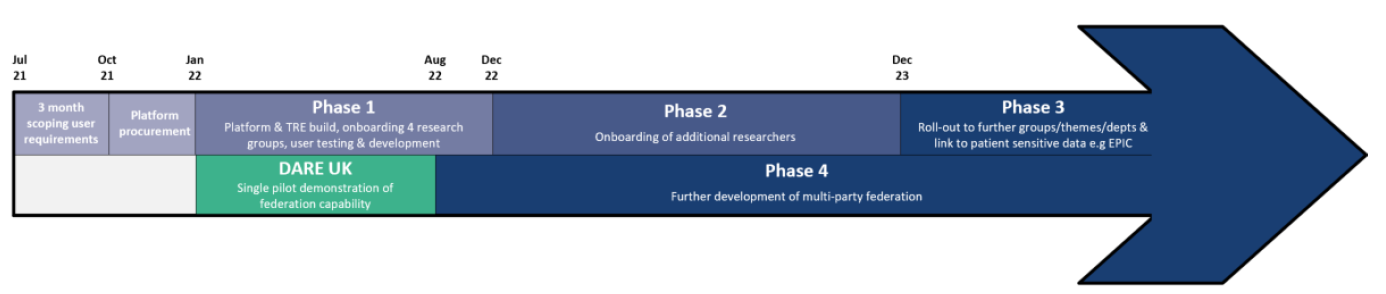 programme timeline.
programme timeline.
What are TREs?
TREs are secure computing environments that hold data, allowing approved researchers to access and analyse information to support scientific studies. You can Read more about TREs.
Who will be the owner of CYNAPSE? Who else is involved in the project?
The University of Cambridge will be the owner of the system. Eastern AHSN and Cambridge University Health Partners have been jointly commissioned to project manage the implementation of the software platform and cloud-based TRE (Lifebit CloudOS) provided by our technology partner Lifebit.
What are the current barriers to others accessing data within the Cambridge Biomedical Campus?
Cambridge-based researchers are already able to ask other researchers for data, based on their experience and knowledge of what is available. This information is then sent in a downloaded format to approved researchers, requiring a lot of computer space and power. CYNAPSE will make it much simpler for approved researchers to access research data safely and securely, without the need for sharing or downloading original datasets.
The CYNAPSE Service Delivery team will ensure any proposed access is safe and appropriate.
What information does the current data contain, and where does it come from?
The data is generated from participants who took part in research studies and contains:
- ‘omic data (for example, DNA, RNA, or protein analysis)
- de-identified phenotypic datasets from previous or current research projects (such as age band, sex, ethnicity)
Some of the data in CYNAPSE were generated through pre-clinical research (this means using human cell lines or animal cells rather than research with human participants).
Does CYNAPSE contain data from patient records?
No. The human data in CYNAPSE comes from large research studies from consenting participants or from pre-clinical research.
However, there are plans to add access to data from patient records through CYNAPSE in future. Where this happens, all information from patients will be combined (known as ‘aggregated’) with all information that could identify an individual removed (known as ‘de-identified’) to protect privacy.
Are you confident the data will be secure from hackers?
We use multiple layers of protection to secure the data from hackers, including:
- All infrastructure resources are placed in a secure private network and access to this network is only accessible through a virtual private network (VPN)
- All infrastructure services, including storage have secure protocols and encryption (can only be read by those with the correct key)
- When data is ‘at rest’ (not currently being accessed or used) encryption and access control is enforced
- Annual ‘penetration testing’ by an independent provider, which involves a simulation of real-world attacks by authorised security professionals in order to find potential weaknesses in security
Will anyone be able to contribute data to CYNAPSE?
Initially, the platform will be tested by a small number of researchers. Once testing is complete, Cambridge-based researchers or clinicians approved by the University of Cambridge Clinical School will be able to contribute research data to CYNAPSE.
Can research teams based outside of Cambridge access or contribute to the database?
CYNAPSE will initially focus on Cambridge-based research teams. Several different specialties and departments are participating in Phase 1 of the programme (building and testing the new platform), with Phase 2 involving additional groups across Cambridge.
However, our long-term vision (Phase 3) is to work with the Genomic Medicine Service Alliance, comprising of Nottingham University Hospitals NHS Trust, University Hospitals of Leicester NHS Trust, and Norfolk and Norwich University Hospitals NHS Foundation Trust.
Who decides what data can be stored in CYNAPSE? Who is responsible for ensuring data has the correct consent?
The data are generated by individual researchers, who are legally responsible for the safe storage and use of the datasets. Any data for inclusion in CYNAPSE will be reviewed to ensure that it meets a minimum set of standards to ensure that it is in a format that is useful to others before it is uploaded. The CYNAPSE Service Delivery
team are in place to develop this process alongside the CYNAPSE patient/public cocreation group and will ensure all Information Governance and data access procedures are followed, check where the data has come from, ensure it has the right approvals and the correct consent in place (or has no need for consent).
Will this add to the data available for systematic review studies?
Not in terms of the actual source data. Any resulting publications & results could be included.
Will researchers be able to access ‘live’ data (data as it is being introduced into the system) or archived data (data that has been stored after permissions have been passed)?
As soon as ‘sharable’ data is uploaded into CYNAPSE, researchers will be able to apply for access to it. Once they have received the relevant approvals, they will be granted access. Only data that has valid permissions will be stored within and uploaded into CYNAPSE. If any relevant permission expires, the data will be disposed of in
accordance with the original requirements of that particular study.
Who is responsible for ensuring only relevant users have access to a dataset?
Only the CYNAPSE Service Delivery team, which includes an Information Governance advisor, will have permissions to grant access to a workspace (and the datasets within it), making sure that researchers have provided details of the proposed research project, alongside NHS Research Ethics (where necessary) and Research &
Development approval.
What happens if a researcher leaves a research group, will they still be able to access data?
If the researcher is no longer employed by the University of Cambridge their access will be revoked. It should be noted that in some cases researchers will move to a visiting worker/collaboration state and retain access.
What are the consequences of misuses of the data?
Researchers accessing data, and often their organisations, sign an agreement that sets out the conditions of use. Breaking these terms could result in legal, financial, reputational, and likely employment consequences.
Will patients be involved in the governance of the programme?
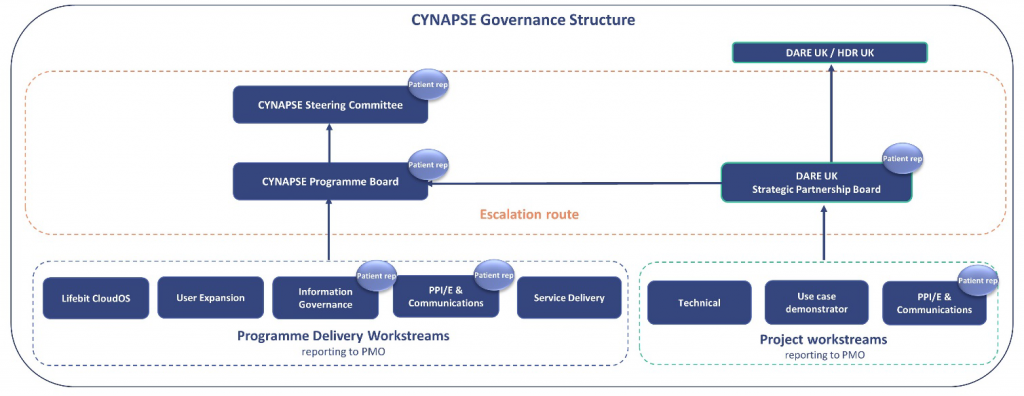
Yes, we have a patient representative on the CYNAPSE Steering Committee, Programme Board, and on a number of workstreams.
Will the partnership with Lifebit last for the whole CYNAPSE programme?
Lifebit’s technology, Lifebit CloudOS, is being used for the new TRE, and Lifebit will be the ones building the federation technology to ‘bridge’ between the TREs for the DARE UK project. A 4-year contract has been signed with Lifebit, after which time the programme will be reviewed before any further contracts are signed.
How are HDR UK involved in CYNAPSE?
HDR UK is not directly associated with the CYNAPSE project, although we aim to incorporate their general recommendations and guidance, for example around the use of TREs. The DARE UK (Data and Analytics Research Environments UK) project is funded by UKRI and delivered in partnership with ADR UK and HDR UK – HDR UK is
responsible for the hands-on project management of phase 1 of the DARE UK programme which runs from January 2022 until August 2022.
Contact:
cynapse@healthinnovationeast.co.uk


