NIHR employees invited to create LGBTQ+ staff network
As part of Pride Month at NIHR, plans have been announced by the NIHR Research Inclusion team to create an NIHR LGBTQ+ Staff Network that will also be open to all NIHR Infrastructure as well as NIHR coordinating centres.
The acronym LGBTQ+ stands for lesbian, gay, bisexual, trans and queer, with the plus encompassing a number of other identities relating to sexual orientation and gender identity including, but not limited to, asexual, pansexual, non-binary, intersex and genderfluid.
NIHR is starting its inclusive staff network journey with an LGBTQ+ Staff Network in response to the interest we received from colleagues during our 2022 Pride activities. The LGBTQ+ Network will serve as a pilot forerunner to other potential NIHR-wide staff networks focusing on different aspects of diversity and inclusion.
Inclusion is one of the NIHR’s operating principles, and embedding inclusion into our systems, processes and culture is one of the areas of strategic focus highlighted in Best Research for Best Health: The Next Chapter.
LGBTQ+ Network membership
The Network will be open to all staff working as part of NIHR (such as staff in coordinating centres, LCRN core teams, delivery staff in LCRN partner organisations and staff in NIHR infrastructure e.g., ARCs and BRCs) who identify as part of the LGBTQ+ community, as well as any staff who consider themselves friends, family or allies to LGBTQ+ people.
Some staff may already be part of LGBTQ+ groups or networks at their employing organisations – the focus of the NIHR LGBTQ+ Staff Network would be to support members around their roles and functions, rather than in relation to their employer.
What is the purpose of the LGBTQ+ Staff Network?
The NIHR Research Inclusion Team is keen to ensure that those within the Network play an active role in deciding what the Network is for.
The NIHR LGBTQ+ Staff Network may provide a space for peer support, developing mentoring relationships and networking across the NIHR for LGBTQ+ staff. The Network may organise events and activities around inclusion of LGBTQ+ people in the work of the NIHR, such as for key dates such as Pride Month, LGBTQ+ History Month, World AIDS Day, Trans Day of Visibility, etc. It may also contribute to activities that support NIHR’s wider strategic inclusion priorities, to ensure inclusivity around sexuality and gender identity in all aspects of the work of the NIHR.
How do we find out more or get involved?
You can get involved in one of two ways:
- register your interest in joining a small working group to help us determine the aims and focus of the LGBTQ+ Staff Network and how it will work
- register your interest in joining the Network once it has been designed.
If you are interested in being part of the Network in either or both of the above capacities, please complete this short form by 14 July 2023 for the NIHR Research Inclusion Team.
Contact:
Please contact the NIHR Research Inclusion Team if you have any further questions or comments.
- Visit the About Us section on our website to find out more about Nurturing Inclusive Research at NIHR Cambridge BRC.
Researchers awarded prestigious Academy of Medical Sciences Fellowships
Four NIHR Cambridge BRC researchers have been elected to the Academy of Medical Sciences Fellowship.
Theme Leads Professors James Rowe and Serena Nik-Zainal, together with researchers Professors Charlotte Coles and Emanuele Di Angelantonio, received the awards in recognition of their outstanding biomedical and health research which has translated into benefits for patients and wider society.



Academy of Medical Sciences President Professor Dame Anne Johnson said: “These new Fellows are pioneering biomedical research and driving life-saving improvements in healthcare. It’s a pleasure to recognise and celebrate their exceptional talent by welcoming them to the Fellowship.”
- This year Fellows were chosen from 353 candidates, and a shortlist of 126 candidates for peer review. To find out more about the Fellowship visit the Academy of Medical Sciences website.
Cambridge researcher wins prestigious award for rare eye disease gene replacement therapy research
Neuroscience researcher Professor Patrick Yu-Wai-Man has won the 2023 Ludwig von Sallmann Clinician-Scientist Award from the Association for Research in Vision and Ophthalmology (ARVO) Foundation.
The award was given in recognition of Prof Yu-Wai-Man’s research on gene replacement therapy for Leber hereditary optic neuropathy (LHON).
LHON is a genetic disorder caused by mutations in mitochondrial DNA (mtDNA). Strictly inherited down the maternal line, it is an important cause of inherited blindness in the young adult population. Currently, there are limited treatment options for LHON and most affected individuals will remain within the legal criteria for blindness.
Prof Yu-Wai-Man said: “I have been working in the field of mitochondrial diseases for nearly 25 years and despite the amazing advances made during that period, finding effective treatments has proven challenging.
“Mitochondrial optic neuropathies have led the way and this award is a recognition of the translational breakthroughs seen in recent years, in particular gene replacement therapy for Leber hereditary optic neuropathy.”
Prof Yu-Wai-Man has worked on several studies using a modified version of the MT-ND4 mitochondrial gene packaged into an adeno-associated viral vector (AAV2) that is injected into the eye. Promising results have been obtained for individuals treated within one year of disease onset with a significant and sustained improvement in vision observed during long-term follow-up.
On receiving the Ludwig von Sallmann Clinician-Scientist Award, Prof Yu-Wai-Man said: “It is a great honour and my hope is that this award will highlight the significant unmet needs for individuals affected with mitochondrial optic neuropathies, which result in significant visual impairment in children and young adults.
“We need to attract more research funding and talents into rare genetic eye diseases. Success breeds success and the future certainly looks bright in this field.”
This article is adapted from arvo.org
Three more great events at this year’s Cambridge Festival – and they’re all from BRC researchers
We’ve already publicised our webinar and family event taking place as part of this year’s Cambridge Festival – now we want to let you know about three more events taking place during the Festival, and they’re all from researchers we fund and support.
Check out the details below – and don’t forget to find out about our events (if you haven’t already) and other NIHR events taking place in Cambridge.
Mental Health
The crisis in mental health in young women and girls: does our education system make it worse? What should we do?
Why are girls and young women suffering levels of stress and anxiety so far in excess of those their mothers and grandmothers experienced? What part does stress at school and university play? Do we have too many exams? Is the content of university courses too onerous? Does the method of assessment at UK universities stress out young women? Why are university students more likely to suffer anxiety than their counterparts at work?
Join Professors Sarah-Jayne Blakemore and Tamsin Ford as they examine some potential solutions, in this hour-long talk from 6-7pm on Monday 27 March, taking place at the Babbage Lecture Theatre, Downing Street.
To find out more and book your place.
Imaging
New medical imaging techniques in the era of A.I.
Dr Joshua Kaggie is an MRI physicist and senior research associate in the Department of Radiology, where he works on developing new imaging techniques. Dr Kaggie moved to Cambridge from Utah in 2015 and has been involved in a range of projects including osteoarthritis, cancer, and dementia imaging.
This talk will discuss some of the more novel imaging techniques that are being developed at Addenbrooke’s Hospital, including the use of heavy hydrogen (deuterium) for use as a new cancer imaging method. This talk will discuss artificial intelligence / machine learning (AI/ML) techniques, their current and future impact on medical imaging and diagnostics, and medical imaging research underway at Addenbrooke’s Hospital – with an emphasis on MRI techniques. The talk will feature a live demonstration of interesting AI developments, some of which may not relate to medicine – yet.
The talk is on Monday, 27 March, from 7.30-8.15pm, at University of Cambridge Admissions Office, New Museums site, Bene’t Street. For more information and to book your place.
Neurodegenerative Disease and Dementias
Memory Matters – an in-person discussion about brain health
This event, taking place from 3.30-5.00pm on Thursday 30 March in the Herchel Smith Building, Robinson Way, will be an opportunity to join researchers for a discussion about brain health in the context of ageing and dementia.
Many of us are worried about our memory as we get older, or following stress to the body (such as long COVID), and it is common to wonder whether we might be developing dementia. But, how much do you know about dementia and how it is detected and treated? In this workshop, you will have the chance to meet, and ask questions of, the doctors, psychologists and nurses who run the world-famous Addenbrooke’s Hospital Memory Clinic.
Short presentations will be followed by a live Q&A session with members of the Memory Clinic team.
For more information and details on how to book.

NICE recommends Cambridge-developed ‘artificial pancreas’ for use on NHS for management of type 1 diabetes
The artificial pancreas for type 1 diabetes – which was developed in Cambridge and supported in trials by the NIHR Cambridge BRC and NIHR Cambridge CRF – could soon be approved for use in the NHS.
The technology is now being recommended by the National Institute for Health and Care Excellence (NICE) as a new way of controlling diabetes and if approved, could be a life-changing tool to manage the disease.
Currently, people with type 1 diabetes rely on multiple, daily finger-prick blood tests and insulin injections to manage their blood sugar, because their pancreas no longer produces insulin.
This new technology combines an insulin pump, continuous glucose monitor and an algorithm to calculate and deliver the right amount of insulin needed. NICE has recommended that the device is offered to patients whose diabetes is difficult to control with other technologies and who are at increased risk of long-term complications – around 105,000 people in England and Wales.
Trialling a new technology
The NIHR Cambridge BRC and CRF have supported the artificial pancreas research from the earliest phases of research more than ten years ago. The device was trialled at the CRF in multiple stages with hundreds of patients, including pregnant women and children.
Research nurses collected blood samples and glucose readings and even conducted overnight studies to understand how effective the device would be to patients whilst they slept. Studies found the artificial pancreas was beneficial in managing people’s diabetes.
Professor Roman Hovorka, who led the team that developed the artificial pancreas, said: “NICE’s recommendations are very welcome and it comes after years of randomised controlled trials (RCTs) at Cambridge.
“These provided the necessary clinical and economic evidence that showed the device has clear health benefits and potential cost savings.
“This technology can literally change lives. If blood glucose levels are too low or too high it can be very damaging and even life-threatening.
“Our trials showed that using the device improved patients’ quality of life and reduced the risk of long-term health complications.”
The closed-loop algorithm developed in Cambridge is now available through CamDiab in 15 countries worldwide, including Australia, France, Italy and Poland.
- See also Artificial pancreas successfully trialled for use by type 2 diabetes patients.
- Artificial pancreas proves ‘life-changing’ for very young children with type 1 diabetes and their families
Gone fishing: highly accurate test for common respiratory viruses uses DNA as ‘bait’
Research supported by NIHR Cambridge BRC has led to a new test that ‘fishes’ for multiple respiratory viruses at once using single strands of DNA as ‘bait’, giving highly accurate results in under an hour.
Cambridge researchers developed the test, which uses DNA ‘nanobait’ to detect the most common respiratory viruses – including influenza, rhinovirus, RSV and COVID-19 – at the same time.
In contrast, PCR (polymerase chain reaction) tests, while highly specific and highly accurate, can only test for a single virus at a time and take several hours to return a result.
While many common respiratory viruses have similar symptoms, they require different treatments. By testing for multiple viruses at once, the researchers say their test will ensure patients get the right treatment quickly and could also reduce the unwarranted use of antibiotics.
In addition, the tests can be used in any setting, and can be easily modified to detect different bacteria and viruses, including potential new variants of SARS-CoV-2, the virus which causes COVID-19. The results are reported in the journal Nature Nanotechnology.
The winter cold, flu and RSV season has arrived in the northern hemisphere, and healthcare workers must make quick decisions about treatment when patients show up in their hospital or clinic.
Similar symptoms, different treatments
“Many respiratory viruses have similar symptoms but require different treatments: we wanted to see if we could search for multiple viruses in parallel,” said Filip Bošković from Cambridge’s Cavendish Laboratory, the paper’s first author. “According to the World Health Organization, respiratory viruses are the cause of death for 20% of children who die under the age of five. If you could come up with a test that could detect multiple viruses quickly and accurately, it could make a huge difference.”
For Bošković, the research is also personal: as a young child, he was in hospital for almost a month with a high fever. Doctors could not figure out the cause of his illness until a PCR machine became available.
“Good diagnostics are the key to good treatments,” said Bošković, who is a PhD student at St John’s College, Cambridge. “People show up at hospital in need of treatment and they might be carrying multiple different viruses, but unless you can discriminate between different viruses, there is a risk patients could receive incorrect treatment.”
PCR tests are powerful, sensitive and accurate, but they require a piece of genome to be copied millions of times, which takes several hours.
The Cambridge researchers wanted to develop a test that uses RNA to detect viruses directly, without the need to copy the genome, but with high enough sensitivity to be useful in a healthcare setting.
“For patients, we know that rapid diagnosis improves their outcome, so being able to detect the infectious agent quickly could save their life,” said co-author Professor Stephen Baker, from the Cambridge Institute of Therapeutic Immunology and Infectious Disease. “For healthcare workers, such a test could be used anywhere, in the UK or in any low- or middle-income setting, which helps ensure patients get the correct treatment quickly and reduce the use of unwarranted antibiotics.”
The researchers based their test on structures built from double strands of DNA with overhanging single strands. These single strands are the ‘bait’: they are programmed to ‘fish’ for specific regions in the RNA of target viruses. The nanobaits are then passed through very tiny holes called nanopores. Nanopore sensing is like a ticker tape reader that transforms molecular structures into digital information in milliseconds. The structure of each nanobait reveals the target virus or its variant.
The researchers showed that the test can easily be reprogrammed to discriminate between viral variants, including variants of the virus that causes COVID-19. The approach enables near 100% specificity due to the precision of the programmable nanobait structures.
“This work elegantly uses new technology to solve multiple current limitations in one go,” said Baker. “One of the things we struggle with most is the rapid and accurate identification of the organisms causing the infection. This technology is a potential game changer; a rapid, low-cost diagnostic platform that is simple and can be used anywhere on any sample.”
A patent on the technology has been filed by Cambridge Enterprise, the University’s commercialisation arm, and co-author Professor Ulrich Keyser has co-founded a company, Cambridge Nucleomics, focused on RNA detection with single-molecule precision.
“Nanobait is based on DNA nanotechnology and will allow for many more exciting applications in the future,” said Keyser, who is based at the Cavendish Laboratory. “For commercial applications and roll-out to the public we will have to convert our nanopore platform into a hand-held device.”
“Bringing together researchers from medicine, physics, engineering and chemistry helped us come up with a truly meaningful solution to a difficult problem,” said Bošković, who received a 2022 PhD award from Cambridge Society for Applied Research for this work.
- The research was supported in part by the European Research Council, the Winton Programme for the Physics of Sustainability, St John’s College, UK Research and Innovation (UKRI), Wellcome and the NIHR Cambridge Biomedical Research Centre.
Cambridge researchers launch study to investigate the impact of neonatal intensive care on premature babies’ sleep patterns and brain development
Cambridge researchers funded by Action Medical Research and the NIHR Cambridge BRC are investigating the impact interruptions in sleep cycles – such as loud noises – have on the development of brain activity in preterm babies in neonatal intensive care (NICU).
The study is significant as preterm babies develop in an environment that is very different from the womb.
The frequent and often painful procedures, bright lights and loud noises in NICU interrupt the natural sleep cycles which are essential for normal brain development – and there is increasing evidence that the different sleep cycles in babies, known as active and quiet sleep, play an important role in the early development of the brain.
Preterm babies to wear ‘swimming caps’ containing sensors to measure brain activity during sleep
In this study, changes in blood flow and oxygen levels will be measured in different parts of the brain using a non-invasive wearable technology, similar to a swimming cap, worn on the head during the quiet and active sleep cycles of very preterm and healthy term infants. This optical imaging technology is called high-density diffuse optical tomography (HD-DOT), and it images functional connections in the brain at the cot-side.
Consultant Neonatologist Professor Topun Austin, who is also Director of the NIHR Cambridge BRC facility the Evelyn Perinatal Imaging Centre, said: “The results from this study could have long-term implications for the care of preterm babies, as this will increase understanding of the sleep states that promote the development of the brain.
“Ultimately this could lead to a traffic light system next to cots, with a red light to indicate a baby is in active or quiet sleep and should not be disturbed, and a green light when the baby is transitioning between sleep states, indicating that the baby can be woken up for tests, feeding or other care.”
In previous research in healthy term infants, Professor Austin and his team showed there was a significant difference in functional connectivity in different sleep states – with babies in active sleep having more connections crossing the brain hemispheres compared with quiet sleep.
This research is needed as in the UK one in every 13 babies are born too soon – before 37 weeks of pregnancy – and around 8,000 babies are born before 32 weeks. Advances in treatment have led to improved survival, however preterm babies have an increased risk of long-term neurodevelopmental complications. Very preterm babies, born before 32 weeks, are at a higher risk of developing behavioural and emotional problems and poor sleep.
- World Prematurity Day takes place each year on 17 November.
- Professor Austin has been interviewed about this research by MIT Technology Review – read the article here.
Off-patent liver disease drug could prevent COVID-19 infection
Cambridge scientists have identified an off-patent drug that can be repurposed to prevent COVID-19 – and may be capable of protecting against future variants of the virus – in research part-funded by the NIHR Cambridge BRC.
The research, published in Nature, showed that an existing drug used to treat a type of liver disease is able to ‘lock’ the doorway by which SARS-CoV-2 enters our cells, a receptor on the cell surface known as ACE2. Because this drug targets the host cells and not the virus, it should protect against future new variants of the virus as well as other coronaviruses that might emerge.
If confirmed in larger clinical trials, this could provide a vital drug for protecting those individuals for whom vaccines are ineffective or inaccessible as well as individuals at increased risk of infection.
Dr Fotios Sampaziotis, from the Wellcome-MRC Cambridge Stem Cell Institute at the University of Cambridge and Addenbrooke’s Hospital, led the research in collaboration with Professor Ludovic Vallier from the Berlin Institute of Health at Charité.
Dr Sampaziotis said: “Vaccines protect us by boosting our immune system so that it can recognise the virus and clear it, or at least weaken it. But vaccines don’t work for everyone – for example patients with a weak immune system – and not everyone have access to them. Also, the virus can mutate to new vaccine-resistant variants.
“We’re interested in finding alternative ways to protect us from SARS-CoV-2 infection that are not dependent on the immune system and could complement vaccination. We’ve discovered a way to close the door to the virus, preventing it from getting into our cells in the first place and protecting us from infection.”
Work on organoids
Dr Sampaziotis had previously been working with organoids – ‘mini-bile ducts’ – to study diseases of the bile ducts. Organoids are clusters of cells that can grow and proliferate in culture, taking on a 3D structure that has the same functions as the part of the organ being studied.
Using these, the researchers found that a molecule known as FXR, which is present in large amounts in these bile duct organoids, directly regulates the viral ‘doorway’ ACE2, effectively opening and closing it. They went on to show that ursodeoxycholic acid (UDCA), an off-patent drug used to treat a form of liver disease known as primary biliary cholangitis, ‘turns down’ FXR and closes the ACE2 doorway.
In this new study, his team showed that they could use the same approach to close the ACE2 doorway in ‘mini-lungs’ and ‘mini-guts’ – representing the two main targets of SARS-CoV-2 – and prevent viral infection.
The next step was to show that the drug could prevent infection not only in lab-grown cells but also in living organisms. For this, they teamed by up with Professor Andrew Owen from the University of Liverpool to show that the drug prevented infection in hamsters exposed to the virus, which are used as the ‘gold-standard’ model for pre-clinical testing of drugs against SARS-CoV-2. Importantly, the hamsters treated with UDCA were protected from the -new at the time- delta variant of the virus, which was new at the time and , which was -at least partially- resistant to existing vaccines.
Professor Owen said: “Although we will need properly-controlled randomised trials to confirm these findings, the data provide compelling evidence that UDCA could work as a drug to protect against COVID-19 and complement vaccination programmes, particularly in vulnerable population groups. As it targets the ACE2 receptor directly, we hope it may be more resilient to changes resulting from the evolution of the SARS-CoV-2 spike, which result in the rapid emergence of new variants.”
Testing using donated lungs not suitable for transplantation
Next, the researchers worked with Professor Andrew Fisher from Newcastle University and Professor Chris Watson from Addenbrooke’s hospital to see if their findings in hamsters held true in human lungs exposed to the virus.
The team took a pair of donated lungs not suitable for transplantation, keeping them breathing outside the body with a ventilator and using a pump to circulate blood-like fluid through them to keep the organs functioning while they could be studied. One lung was given the drug, but both were exposed to SARS-CoV-2. Sure enough, the lung that received the drug did not become infected, while the other lung did.
Professor Fisher said: “This is one of the first studies to test the effect of a drug in a whole human organ while it’s being perfused. This could prove important for organ transplantation – given the risks of passing on COVID-19 through transplanted organs, it could open up the possibility of treating organs with drugs to clear the virus before transplantation.”
Testing on healthy volunteers
Moving next to human volunteers, the Cambridge team collaborated with Professor Ansgar Lohse from the University Medical Centre Hamburg-Eppendorf in Germany.
Professor Lohse explained: “We recruited eight healthy volunteers to receive the drug. When we swabbed the noses of these volunteers, we found lower levels of ACE2, suggesting that the virus would have fewer opportunities to break into and infect their nasal cells – the main gateway for the virus.”.
While it wasn’t possible to run a full-scale clinical trial, the researchers did the next best thing: looking at data on COVID-19 outcomes from two independent cohorts of patients, comparing those individuals who were already taking UDCA for their liver conditions against patients not receiving the drug. They found that patients receiving UDCA were less likely to develop severe COVID-19 and be hospitalised.
A safe, affordable variant-proof drug
First author and PhD candidate Teresa Brevini from the University of Cambridge said: “This unique study gave us the opportunity to do really translational science, using a laboratory finding to directly address a clinical need.
“Using almost every approach at our fingertips we showed that an existing drug shuts the door on the virus and can protect us from COVID-19. Importantly, because this drug works on our cells, it is not affected by mutations in the virus and should be effective even as new variants emerge.”
Dr Sampaziotis said the drug could be an affordable and effective way of protecting those for whom the COVID-19 vaccine is ineffective or inaccessible. “We have used UDCA in clinic for many years, so we know it’s safe and very well tolerated, which makes administering it to individuals with high COVID-19 risk straightforward.
“This tablet costs little, can be produced in large quantities fast and easily stored or shipped, which makes it easy to rapidly deploy during outbreaks – especially against vaccine-resistant variants, when it might be the only line of protection while waiting for new vaccines to be developed. We are optimistic that this drug could become an important weapon in our fight against COVID-19.”
The research was largely funded by UK Research & Innovation, the European Association for the Study of the Liver, the NIHR Cambridge Biomedical Research Centre and the Evelyn Trust.
- Watch a video of Dr Sampaziotis explaining the implications of this research:
Cambridge researchers develop safe, affordable device for prostate cancer diagnosis
A new medical device developed at Addenbrooke’s and supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility aims to reduce the risk of infection in prostate patients – and save time and money.
The device – called the Cambridge Prostate Biopsy Device (CamPROBE) – has been developed by urology specialist Professor Vincent Gnanapragasam and his team at Cambridge University Hospitals and Cambridge University.
Traditionally, prostate patients have had transrectal biopsies, where a sample of tissue is removed from the prostate using a thin needle that is inserted through the rectum and into the prostate.
This carries a significant risk of side effects, including urinary infections and severe sepsis -and medical and professional bodies now advocate using instead the transperineal route, which is the space between the legs and under the scrotum.
The CamPROBE uses the transperinal route – making it safer for patients. It’s also cost-effective and simple to use – the procedure can be carried out in outpatients under local anaesthetic.
Urology consultant Professor Gnanapragasam (pictured below) said: “In trials cancer detection rates were equivalent to other means of biopsy.
“Procedure times were short and only low amounts of local anaesthetic were required, yet low pain scores were reported by patients.

“More than 85% of patients said they would recommend the CamPROBE procedure as a method of having a prostate biopsy done.”
The CamPROBE aims to make the lives of patients better through a simple and low pain approach of prostate cancer detection, hopefully benefitting the millions of men who have prostate biopsies every year.
A licensing agreement for CamPROBE has been agreed with product development company JEB Technologies.
NIHR statement on the death of Her Majesty Queen Elizabeth II
We at the NIHR offer our sincerest condolences to the royal family on the passing of Her Majesty Queen Elizabeth II. We share in the grief of the whole nation at the death of our remarkable monarch, and reflect with great pride upon her incredible reign.
Research reveals how genetic mutations cause kidney cancer
Researchers at the University of Cambridge have shown that genetic mutations associated with kidney cancer rely on factors that regulate normal kidney cells in order to develop into cancer cells.
The study suggests that similar mechanisms could explain why cancer mutations cause specific types of cancer to develop.
Inherited genetic variants and mutations that are randomly acquired during the lifetime of an individual can lead to the development of cancer. But different mutations tend to cause different types of cancer.
While this cancer-specificity of mutations has been clear for decades, exactly why mutations can lead to the formation of tumours in some tissues but not others has remained poorly understood.
All tissues have specific functions that are ultimately dependent on the instructions encoded by the genome. These instructions are read by a group of proteins called transcription factors that recognise specific DNA sequences and ensure that the right regions of the genome are active in each cell. This mechanism allows the same genome to control the functions of diverse cell types with vastly different characteristics.
In this study, published in the journal Nature, the researchers tested whether the transcription factors that control tissue-specific functions of normal kidney cells were also required for the growth of kidney cancers.
They used a combination of advanced genomic tools, experimental cancer models and analysis of large human data sets.
The results show that the ability of kidney cancer-associated genetic alterations to promote tumour formation was dependent on transcription factors that are specifically active in normal kidney cells.
When kidney-specific transcription factors were inactivated experimentally, the cancer mutations were no longer capable of activating genes that are important for tumour growth.
Senior author Dr Sakari Vanharanta said: “Our results provide some insight into the molecular mechanisms that dictate the cancer type-specificity of mutations.
“Genetic alterations that cause kidney cancer rely on factors that under normal conditions regulate specific functions of healthy kidney cells. If these factors are not present, as they are not in most other cell types, the process that eventually leads to cancer formation does not proceed.”
Large genetic alterations commonly observed in advanced, metastatic kidney cancer cells also relied on kidney-specific transcription factors for their cancer-promoting effect.
The common genetic variant characterised in this study is carried by the majority of individuals of European descent and it increases the risk of kidney cancer. Overall, the molecular mechanisms described in this work are likely to be important for a large proportion of kidney cancers.
Professor Grant Stewart, study co-author and co-lead of the CRUK Cambridge Centre Urological Malignancies Programme, said: “[This] paves the way to new thinking on how to develop new treatments for kidney cancer and even prevent it from developing in the first place.
“These same mechanisms might also be at play in other cancer types, making this study highly relevant across all cancers”.
Cambridge researchers to receive nearly £4m to tackle cancer roadblocks
NIHR Cambridge BRC researchers are among the Cambridge scientists to receive £3,938,500 as part of Cancer Grand Challenges, a major initiative co-founded by Cancer Research UK and the National Cancer Institute in the US, which aims to encourage the world’s leading cancer researchers to take on some of the toughest challenges in cancer research.
The eDyNAmiC (extrachromosomal DNA in Cancer) team will investigate new ways to combat treatment resistant cancers, while the CANCAN (CANcer Cachexia Action Network) team hopes to prevent cachexia, where patients ‘waste away’ in the later stages of their disease.
Funding for the Cambridge-based projects is part of an overall £80 million awarded this week to four elite global teams who will deepen our understanding of cancer through international collaboration leading to new advances for people with cancer.
Extrachromosomal DNA – present in up to a third of all tumours, helps to evade treatment
The Stanford University-led eDyNAmiC team, which includes Professor Serena Nik Zainal at the University of Cambridge, hopes to tackle tumour evolution, which is driven by circular pieces of tumour DNA which exist outside the tumour and pose a major problem by enabling tumours to resist treatment.
Research is now revealing that a major driver of tumour evolution is extrachromosomal DNA (ecDNA). These small circular DNA particles enable cells to rapidly change their genomes and so evade the immune system.
EcDNA doesn’t follow the rules of normal chromosomes, providing tumours a way to evolve and change their genomes to evade treatment.
Although first observed in cancer in 1965, researchers are only beginning to understand the extent to which it is prevalent in around a third of cancers and how it helps tumours to become more resistant, aggressive and affect patient survival.
The goal of eDyNAmiC is to understand how extrachromosomal DNA is created, to find vulnerabilities and then to develop new ways to target these in some of the hardest cancers to treat, including glioblastoma, lung and oesophageal cancer.
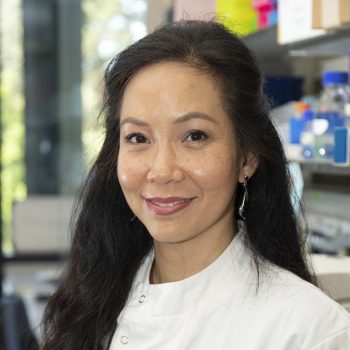
Co-investigator Professor Nik-Zainal (pictured, below), of the University of Cambridge Early Cancer Institute and Department of Medical Genetics, said: “My team and I are so excited to be part of this collaboration studying this phenomenon of extra pieces of DNA called ecDNA.
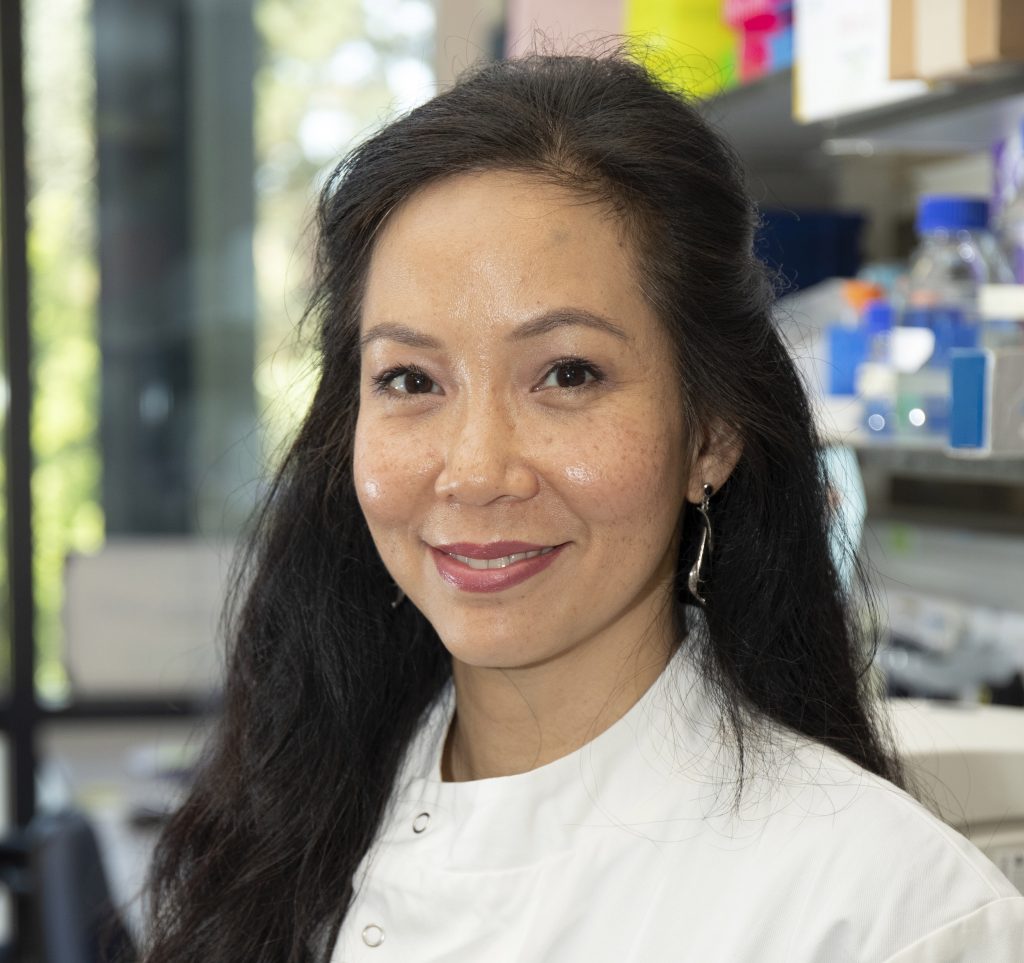
“What a privilege it is to be entrusted to explore how ecDNAs cause cancer and drive them to be aggressive. We hope that what we learn will bring real benefits to cancer patients in due course.”
Research aims to improve quality of life for patients with cancer cachexia
The CANCAN team is led by US researchers who will work with co-investigators Professor Sir Stephen O’Rahilly and Dr Tony Coll, of the Wellcome-MRC Institute of Metabolic Science, and Dr Giulia Biffi, of the CRUK Cambridge Institute, to explore the underpinning mechanisms behind cancer cachexia – a debilitating wasting condition many people experience in the later stages of the disease.

Cachexia syndrome is characterised by poor appetite and extensive weight loss from both skeletal muscle and fatty tissue and is still not fully understood.
It is hoped further research can help develop new treatments to improve quality of life for cancer patients and set the standard for cachexia management around the world.
Professor Sir Stephen O’Rahilly said: “For many decades we have studied how a range of hormones act on the brain to regulate appetite and body weight.
“Many of the insights that we have gained through our previous research in obesity are likely to be highly relevant to cancer cachexia, a condition where hormonal and metabolic changes secondary to cancer impact on the brain to reduce, rather than increase appetite.
“We have recently discovered a pathway in the brain which is key to controlling whether we put food calories in excess of our basic needs into fat or into muscle. This pathway is likely to be highly relevant to patients with cancer cachexia who are particularly affected by a loss of muscle.”
Dr Giulia Biffi’s research focuses on understanding the biology of the tumour microenvironment in pancreatic cancer with the aim of developing new treatments and diagnostics.
Dr Biffi, who co-leads the CRUK Cambridge Centre Pancreatic Cancer Programme, said: “Cachexia is predominant in pancreatic cancer patients; it increases patient mortality and can prevent patients accessing treatment as they are too weak. If we can identify ways to treat cachexia more people could be treated for their cancer.
“This is a fantastic opportunity to interact with a highly multi-disciplinary team to bring our different scientific and clinical strategies towards a single common goal.”
- Dr Coll with Dr Claire Connell have recently opened a clinical study at Addenbrooke’s hospital which aims to understand the mechanisms underlying weight loss in cancer patients by investigating changes to metabolism and the immune system. They hope the findings from the Metabolic and Immunological Phenotyping in Patients with Cancer (MIPPaC) study will guide future research and help to design treatments that can alleviate or prevent weight loss and improve outcomes for cancer patients.
Consensus opens door to worldwide improvements in breast cancer radiotherapy treatment
A panel of European experts and patients has identified a way to achieve major changes in the way radiotherapy treatment for breast cancer patients is delivered around the world.
The Consensus from ESTRO–ACROP: European Society for Radiotherapy and Oncology – Advisory Committee in Radiation Oncology Practice, which has been published online by Lancet Oncology, states that 3-week breast radiotherapy can be offered to any patient for any indication and 1-week breast radiotherapy can be offered for patients who do not need lymph node radiotherapy.
It follows months of work from the panel. They first looked at research on increased doses of radiation over a shorter timeframe (called hypofractionation), and then compiled consensus statements.
These were reviewed by experts from hospitals across Europe, Brazil and Israel, who graded how much they either agreed or disagreed with the statements.
Finally, the panel selected the statements that scored the highest and agreed a formal Consensus.
Professor Charlotte Coles, who is also an NIHR Research Professor, Deputy Head of the Department of Oncology at the University of Cambridge and a member of the core ESTRO-ACROP consensus group, said: “In the clinical world Consensus statements can drive change – so this really is the next step to changing standard of care in Europe and the rest of the world.
“We knew the evidence was there to support hypofractionation – there have been some excellent clinical trials over the last couple of decades. These include the recent UK FAST-Forward trial that showed that 1-week breast radiotherapy is at least as good as conventional treatment in preventing cancer returning with similar side effects, but has far fewer treatment visits for patients.”
Hypofractionation is already standard of care in the UK, but it has been much slower to implement elsewhere. In addition, there are huge areas of the world, for example in parts of Africa, where access to radiotherapy treatment is extremely poor.
Professor Coles said: “Changing from three or even five weeks in some places to one week will make breast radiotherapy treatment a realistic goal for everyone who needs it.
“There are other benefits too, including less travel for treatment, less time off work, less time in recovery.
“What the Consensus is saying is that doctors could treat three breast cancer patients in the time that it used to take to treat just one, which would mean more patients could access breast radiotherapy.
“The good things about reducing the number of weeks is we’re not giving less treatment, it’s the same treatment only over one week, in more concentrated doses.
“The science behind it is based on how breast cancer reacts to hypofractionation, and it’s a happy consequence that it’s also quicker and cheaper.”
Professor Coles is delighted that the Consensus will be published in Lancet Oncology: “This is the first time that a consensus or guideline from ESTRO-ACROP has been published in a high-impact journal like Lancet Oncology.
“This will help us enormously in our work to influence policy makers to ensure equity of access to evidence-based radiotherapy.
“We know Consensus statements drive change but in many places health care providers are still reimbursed for each dose they deliver and not for the whole course. This is a disincentive for providing evidence-based hypofractionation and needs to be changed.
“We want equity so that everyone can access the radiotherapy they need and for providers to be incentivised by the quality of the radiotherapy they deliver rather than the number of treatments so that the patient experience is better.
“I am currently chairing The Lancet Breast Cancer Commission, which will look at reimbursement as part of its remit. We hope this Consensus will convince policy-makers of the health and economic value of breast hypofractionation.
“The ultimate goal is to make high-quality breast cancer radiotherapy accessible for everyone, no matter where they live.
“We’ve already changed practice in the UK, we now need to change it internationally.”
- “European Society for Radiotherapy and consensus recommendations on patient selection and dose and fractionation for external beam radiation therapy in early breast cancer” will appear in print in Lancet Oncology in January 2022.
PhD Programme for Health Professionals now open for applications
An exciting PhD Programme for Health Professionals has been established in partnership between the Universities of Cambridge and East Anglia and the Wellcome Sanger Institute. From 2022, funding is available for seven Fellowships each year for the next five years.
The scheme is ideally suited to clinical health professionals of the highest calibre, seeking to pursue a successful academic career.
If appropriate, predoctoral research placements in different research groups enable fellows to make an informed choice of supervisor and project, leading to a PhD.
In addition, the programme offers bespoke training and mentoring for a successful research experience, and guidance postdoctorally. Many graduates of the previous Clinical PhD Programme have continued in an academic pathway, gaining higher research fellowships, and achieving academic faculty positions.
Fellowships cover the broad themes of Genetic and Molecular Basis of Disease; Pathogens, Infectious Disease and Immunity; Physiology, Pathophysiology and Experimental Medicine; Behaviour, Mental Health and Neurological Diseases; Epidemiology, Prevention and Public Health; Health Care Delivery and Improvement.
Application deadline: Monday 17 January 2022 (midday UK time).
Visit the Wellcome-MRC Institute of Metabolic Science website for further details or contact the Programme Administrator with any queries.
Key mutations in Alpha variant enable SARS-CoV-2 to overcome evolutionary weak points
One of the key mutations seen in the ‘Alpha variant’ of SARS-CoV-2 – the deletion of two amino acids, H69/V70 – enables the virus to overcome chinks in its armour as it evolves, say an international team of scientists.
SARS-CoV-2 is a coronavirus, so named because spike proteins on its surface give it the appearance of a crown (‘corona’). The spike proteins bind to ACE2, a protein receptor found on the surface of cells in our body. Both the spike protein and ACE2 are then cleaved, allowing genetic material from the virus to enter the host cell. The virus manipulates the host cell’s machinery to allow the virus to replicate and spread.
As SARS-CoV-2 divides and replicates, errors in its genetic makeup cause it to mutate. Some mutations make the virus more transmissible or more infectious, some help it evade the immune response, potentially making vaccines less effective, while others have little effect.
Towards the end of 2020, Cambridge scientists observed SARS-CoV-2 mutating in the case of an immunocompromised patient treated with convalescent plasma. In particular, they saw the emergence of a key mutation – the deletion of two amino acids, H69/V70, in the spike protein. This deletion was later found in B1.1.7, the variant that led to the UK being forced once again into strict lockdown in December (now referred to as the ‘Alpha variant’).
Now, in research published in the journal Cell Reports, researchers show that the deletion H69/V70 is present in more than 600,000 SARS-CoV-2 genome sequences worldwide, and has seen global expansion, particularly across much of Europe, Africa and Asia.
The research was led by scientists at the University of Cambridge, MRC-University of Glasgow Centre for Virus Research, The Pirbright Institute, MRC Laboratory of Molecular Biology, and Vir Biotechnology.
Professor Ravi Gupta from the Cambridge Institute of Therapeutic Immunology and Infectious Disease at the University of Cambridge, the study’s senior author, said: “Although we first saw this mutation in an immunocompromised patient and then in the Kent – now ‘Alpha’ – variant, when we looked at samples from around the world, we saw that this mutation has occurred and spread multiple times independently.”
Working under secure conditions, Professor Gupta and colleagues used a ‘pseudotype virus’ – a harmless virus that displays SARS-CoV-2 spike proteins with the H69/V70 deletion – to understand how the spike protein interacts with host cells and what makes this mutation so important.
When they tested this virus against blood sera taken from fifteen individuals who had recovered from infection, they found that the deletion did not allow the virus to ‘escape’ neutralising antibodies made after being vaccinated or after previous infection. Instead, the team found that the deletion makes the virus twice as infective – that is, at breaking into the host’s cells – as a virus that dominated global infections during the latter half of 2020. This was because virus particles carrying the deletion had a greater number of mature spike proteins on their surface. This allows the virus to then replicate efficiently even when it has other mutations that might otherwise hinder the virus.
“When viruses replicate, any mutations they acquire can act as a double-edged sword: a mutation that enables the virus to evade the immune system might, for example, affect how well it is able to replicate,” said Professor Gupta.
“What we saw with the H69/V70 deletion was that in some cases, the deletion helped the virus compensate for the negative effects that came with other mutations which allowed the virus to escape the immune response. In other words, the deletion allowed these variants to have their cake and eat it – they were both better at escaping immunity and more infectious.”
Dr Dalan Bailey from The Pirbright Institute, who co-led the research, added: “In evolutionary terms, when a virus develops a weakness, it can lead to its demise, but the H69/V70 deletion means that the virus is able to mutate further than it otherwise would. This is likely to explain why these deletions are now so widespread.”
Bo Meng from the Department of Medicine at the University of Cambridge, first author on the paper, said: “Understanding the significance of key mutations is important because it enables us to predict how a new variant might behave in humans when it is first identified. This means we can implement public health and containment strategies early on.”
The research was supported by Wellcome, the Medical Research Council, the Bill & Melinda Gates Foundation and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre.
- The paper Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the variant of concern lineage B.1.1.7 is available online in the open-access journal Cell Reports.
Upgrading PPE for staff working on COVID-19 wards cut hospital-acquired infections dramatically
When Addenbrooke’s Hospital in Cambridge upgraded its face masks for staff working on COVID-19 wards to filtering face piece 3 (FFP3) respirators, it saw a dramatic fall – up to 100% – in hospital-acquired SARS-CoV-2 infections among these staff.
The findings are reported by a team at the University of Cambridge and Cambridge University Hospitals (CUH) NHS Foundation Trust. The research has not yet been peer-reviewed, but is being released early because of the urgent need to share information relating to the pandemic.
Until recently, advice from Public Health England recommended that healthcare workers caring for patients with COVID-19 should use fluid resistant surgical masks type IIR (FRSMs) as respiratory protective equipment; if aerosol-generating procedures were being carried out (for example inserting a breathing tube into the patient’s windpipe), then the guidance recommended the use of an FFP3 respirator. PHE has recently updated its guidance to oblige NHS organisations to assess the risk that COVID-19 poses to staff and provide FFP3 respirators where appropriate.
Since the start of the pandemic, CUH has been screening its healthcare workers regularly for SARS-CoV-2, even where they show no symptoms. They found that healthcare workers caring for patients with COVID-19 were at a greater risk of infection than staff on non-COVID-19 wards, even when using the recommended respiratory protective equipment. As a result, its infection control committee implemented a change in respiratory protective equipment for staff on COVID-19 wards, from FRSMs to FFP3 respirators.
Prior to the change in respiratory protective equipment, cases were higher on COVID-19 wards compared with non-COVID-19 wards in seven out of the eight weeks analysed by the team. Following the change in protective equipment, the incidence of infection on the two types of ward was similar.
The results suggest that prior to the change, almost all cases among healthcare workers on non-COVID-19 wards were caused by community-acquired infection, whereas cases among healthcare workers on COVID-19 wards were caused by both community-acquired infection and direct, ward-based infection from patients with COVID-19 – but that these direct infections were effectively mitigated by the use of FFP3 respirators.
To calculate the risk of infection for healthcare workers working on COVID-19 and non-COVID-19 wards, the researchers developed a simple mathematical model.
Dr Mark Ferris from the University of Cambridge’s Occupational Health Service, one of the study’s authors, said: “Healthcare workers – particularly those working on COVID-19 wards – are much more likely to be exposed to coronavirus, so it’s important we understand the best ways of keeping them safe.
“Based on data collected during the second wave of the SARS-CoV-2 pandemic in the UK, we developed a mathematical model to look at the risks faced by those staff dealing with COVID-19 patients on a day to day basis. This showed us the huge effect that using better PPE could have in reducing the risk to healthcare workers.”
According to their model, the risk of direct infection from working on a non-COVID-19 ward was low throughout the study period, and consistently lower than the risk of community-based exposure.
By contrast, the risk of direct infection from working on a COVID-19 ward before the change in respiratory protective equipment was considerably higher than the risk of community-based exposure: staff on COVID-19 wards were at 47 times greater risk of acquiring infection while on the ward than staff working on a non-COVID-19 ward.
Crucially, however, the model showed that the introduction of FFP3 respirators provided up to 100% protection against direct, ward-based COVID-19 infection.
Dr Chris Illingworth from the MRC Biostatistics Unit at the University of Cambridge, said: “Before the face masks were upgraded, the majority of infections among healthcare workers on the COVID-19 wards were likely due to direct exposure to patients with COVID-19.
“Once FFP3 respirators were introduced, the number of cases attributed to exposure on COVID-19 wards dropped dramatically – in fact, our model suggests that FFP3 respirators may have cut ward-based infection to zero.”
Dr Nicholas Matheson from the Department of Medicine at the University of Cambridge, said: “Although more research will be needed to confirm our findings, we recommend that, in accordance with the precautionary principle, guidelines for respiratory protective equipment are further revised until more definitive information is available.”
Dr Michael Weekes from the Department of Medicine at the University of Cambridge, added: “Our data suggest there’s an urgent need to look at the PPE offered to healthcare workers on the frontline. Upgrading the equipment so that FFP3 masks are offered to all healthcare workers caring for patients with COVID-19 could reduce the number of infections, keep more hospital staff safe and remove some of the burden on already stretched healthcare services caused by absence of key staff due to illness. Vaccination is clearly also an absolute priority for anyone who hasn’t yet taken up their offer.”
The research was funded by Wellcome, the Addenbrooke’s Charitable Trust, UK Research and Innovation, and the NIHR Cambridge Biomedical Research Centre.
- For a copy of the paper see: FFP3 respirators protect healthcare workers against infection with SARS-CoV-2.
Cambridge researchers win government funding for their artificial intelligence (AI) technologies
Technologies developed by Cambridge researchers that use artificial intelligence to speed up diagnosis and improve patient care have been successful in the latest round of the £140million Artificial Intelligence in Health and Care Award.
In total four AI projects, which Cambridge researchers either led or collaborated in, received funding, which was announced by Secretary of State for Health, Matt Hancock, on 16 June.
It means that the researchers will be able to take the technology one step closer to being used in the NHS to benefit patients.
AI-systems for improving blood transfusion outcomes
Professor Emanuele Di Angelantonio from the NIHR Blood and Transplant Research Unit in Donor Health and Genomics, Dr William Astle NHSBT Senior Lecturer in Statistical Science in the MRC Biostatistics Unit and their collaborators at UCLH, Oxford University and international blood services, have been awarded more than £1million to improve, develop and implement AI-systems for genetic blood group typing, the automated stocking of blood according to type and the precision matching of patients to blood units.
Professor di Angelantonio said: “There are over thirty known blood group systems and each system can create different blood types.
“For patients receiving regular blood transfusions, it’s vital that the blood they receive is compatible with their own. If the blood is not matched, it can cause complications which can get worse over time – but at the moment it costs the NHS a lot of time and money to run the tests that are needed to measure a complete set of blood groups.
“It is now very cheap to measure blood groups genetically – we hope to blood type 100,000 blood donors and 500 sickle cell disease patients genetically in 2021. This will allow us to use AI to match sickle cell disease patients – who stand to benefit hugely from more precise blood matching – to donors.”
Dr Astle added: “This NIHR AI Award funding will help us to extend and develop the AI methods required for genetic blood matching, which should reduce the avoidable harm caused by transfusion reactions in the NHS.
“This has the potential to transform the quality of clinical care for patients dependent on transfusion, who can become untransfusable if they regularly receive mismatched blood.”
AI-enabled spine fracture pathway
In this multi-centre study, five hospitals including Cambridge University Hospitals are using an AI solution to detect osteoporotic fractures and identify new patients for treatment.
Dr Ken Poole, who is leading the study in Cambridge, said: “Osteoporosis is a common bone disease. Breaking a bone in the spine is a clear sign of it, but many patients don’t even realise they have a spine fracture at the time. They mistake the symptoms for ordinary back pain, and ignoring that cue can then lead to a cascade of more spine fractures or even a hip fracture, with devastating consequences.
“Each year over two million people undergo scans that include the spine, for various reasons such as lung or bowel problems. Remarkably, up to one in 20 of people having these scans could have spine fracture, although very few fractures are recognised or acted upon.
“This project will use an innovative AI software that automatically looks at existing CT scans to find these fractures and brings them directly to the specialist team’s attention, to see if the patient needs bone-strengthening lifestyle advice and medicines.
“We believe that this ‘AI-enabled spine fracture pathway’ will improve patient health and reduce costs to hospitals – and ensure tens of thousands of adults with undetected spine fractures are identified and protected against having future potentially life-changing fractures.”
This project is being run with Zebra-Med, an AI and machine learning company headquartered in Israel.
AI to differentiate tumour and healthy tissue on cancer scans
Consultant oncologist Dr Raj Jena has received an AI Award for 12 months to accelerate the process of registering AI technology that can spot the difference between tumours and healthy tissue on cancer scans as a medical device.
Once registered, it can then begin to be rolled out across key NIHR BRCs.
The NIHR Cambridge and Birmingham BRCs are using open-source AI tools from Microsoft Project InnerEye to differentiate tumour from healthy tissue on cancer scans. This is called ‘segmenting’ and takes place prior to radiotherapy treatment.
Dr Jena said: “This saves clinicians’ time, and reduces the time between the scan and commencing radiotherapy treatment.”
Monitoring slow-growing brain tumours
Certain types of brain tumour are deemed low-risk, as they grow so slowly. This project, led by Hon. Consultant Neurosurgeon Stephen Price, aims to develop AI to measure the volume of tumours from scans, and learn which are at risk of growth, to ensure those patients are monitored more frequently, and others can be reassured that their tumour is lower risk.
Professor Miles Parkes, Director of the NIHR Cambridge BRC, said: “This is fantastic news and congratulations to everyone involved.
“The Award will help increase the impact of the AI-driven technologies that Ken, Emanuele, Will, Raj and Stephen are developing.
“Their innovations will not only offer faster and more personalised diagnosis, they will also provide the evidence we need to demonstrate the effectiveness and safety of AI-driven technologies in health and social care.
“The technologies our researchers are developing today have the potential to transform clinical care for thousands of patients across the country.”
Patient and Public Involvement and Engagement (PPI/E) Strategy Lead Dr Amanda Stranks said: “For these technologies to be successful in the clinic, it is essential that patients are at the heart of their design and usage.
“It’s also great to see the NIHR asking for clear evidence of strong patient and public involvement strategies in the projects they are funding.
“For example, the AI osteoporosis study under Ken Poole in Cambridge will have patients represented on the study steering committee, project management team and locally, to ensure the project remains patient-centred throughout its life cycle.
“The AI system for improving blood transfusion outcomes also has actively involved patients, including setting up a patient panel to advise on the project.”
- All the AI projects that receive awards will be independently evaluated for their effectiveness, safety and value in the NHS settings in which they are deployed. This will add to the evidence base and inform the onward adoption and scaling of the technology.
Updated risk model helps doctors predict and prevent cardiovascular disease
Research developing the European Society of Cardiology (ESC) cardiovascular disease (CVD) risk prediction calculator to aid efforts to reduce the burden of CVD in Europe has been published today by European Heart Journal.
The research – carried out by the SCORE2 Working Group and the ESC Cardiovascular Risk Collaboration – analysed data from nearly 700,000 mainly middle-aged participants in 45 large-scale studies to develop risk prediction models (SCORE2) tailored for use in European countries.
The participants did not have previous history of CVD at the outset and 30,000 had a CVD event (heart attack or stroke) during the first 10 years of follow up.
These risk models were then statistically adapted or ‘recalibrated’ to more accurately estimate CVD risk for contemporary populations in four European risk regions, using data on population-specific CVD incidence rates and risk factor values from 10.8 million individuals.
SCORE2 will replace the original SCORE (Systematic COronary Risk Evaluation) model, and is adopted by the upcoming European Guidelines on CVD Prevention in Clinical Practice.

Professor Di Angelantonio (pictured), who with Dr Lisa Pennells and Dr Stephen Kaptoge led the research from the University of Cambridge, said: “The original research focused solely on predicting and preventing mortality, using data from the 1980s.
“But now more people survive heart attacks and strokes than die from them, especially younger people, so we wanted to show the absolute risk scores of people having non-fatal as well as fatal CVD within 10 years.
“This new highly collaborative effort was developed using data from dozens of countries, including exceptionally powerful, extensive and complementary datasets of contemporary relevance to European populations.
“As this risk prediction tool is superior to its predecessors, it should have substantial real-world impact by improving the primary prevention of cardiovascular disease across Europe through helping doctors to identify high-risk patients who may benefit from lifestyle change or preventative medication.”
- SCORE2 risk prediction algorithms: revised models to estimate 10-year risk of cardiovascular disease in Europe was published by the European Heart Journal on 14 June 2021.
Join us in supporting International Clinical Trials Day
Every year 20 May is celebrated by healthcare workers, researchers, patients and communities around the world to mark International Clinical Trials Day.
On this day in 1747, ship surgeon James Lind started what is believed to be the world’s first randomised clinical trial, when he asked 12 sailors who all had scurvy to ingest six different treatments including cider, vinegar, sea water and an orange and lemon. Within days the man who had received the citrus fruit had recovered so much that he was appointed nurse to the others on the trial.
Make future research breakthroughs possible
To coincide with International Clinical Trials Day, the NIHR is running its Be Part of Research campaign.
This celebrates the thousands of people who volunteered to be part of research including taking part in the fight back against COVID-19. At the same time, the campaign encourages more people to take part in research to ensure future medical breakthroughs happen.
Being part of research: what our patients and volunteers say
- Volunteers are still needed for studies into all conditions, including diabetes, cancer, mental health and dementia: find out what research is taking place near you.
- Do you use Twitter? Include the #BePartofResearch hashtag and help promote the value of clinical research. Don’t forget to follow us on Twitter for more patient quotes about taking part in research!
Save your energy! New tool shows algorithms’ environmental impact
Data science and artificial intelligence are transforming UK healthcare – but at an environmental cost.
The data centres housing the supercomputers that run the algorithms account for 100 megatonnes of CO2 emissions every year – roughly the same as US commercial aviation. It takes more and more energy to run increasingly complex algorithms – and even a best-case scenario predicts a three-fold increase in the sector’s energy needs by 2030.
Now Cambridge scientists supported by the NIHR Cambridge BRC have developed a freely available tool which allows anyone to estimate the carbon footprint of their computations.
Dr Michael Inouye from the University of Cambridge, who supervised the development of Green Algorithms, said: “We’ve been using computers, supercomputers and cloud computation for many years now, yet there hasn’t been an easy way to work out how much greenhouse gases our computations emit.
“High-performance and large-scale computing have led to many scientific breakthroughs; for example, in astrophysics we now know what black holes 55 million light-years away look like, and in genomics, we now know of thousands of genetic variants for common diseases. We didn’t know the environmental impact of using data science to uncover these discoveries, nor did we have the tools to estimate what the next-generation of breakthroughs would take in terms of carbon footprint.
“So our team set about developing a free and easy-to-use online tool, Green Algorithms, for users to estimate the carbon footprint of any computational task.”
Know your computation’s carbon footprint
Want to know what the weather will be like tomorrow in your home town? A supercomputer in Reading will do that for you – at an environmental cost.
Using Green Algorithms, Dr Inouye’s team calculated that the computations it runs for just one day’s forecasts are equivalent in terms of greenhouse gas (GHG) emissions to driving 1,708 km or taking three return flights between Paris and London. It would take a mature tree over 27 years to sequester this amount of carbon.
Dr Inouye said: “Green Algorithms takes into account what’s ‘under the hood’ of a computer, the length of time it’s used and the location of the resources it utilises to estimate the energy used and the kilograms of CO2 emitted.
“It then shows the equivalent footprint in car and plane journeys, and ‘tree months’, or the number of months a mature tree needs to absorb a given quantity of carbon.
“Green Algorithms also makes specific recommendations for reducing computing’s carbon footprint. For example do you really want to continue using data centres which are heavily reliant on coal for power, even if they have faster processors? What would be the carbon savings or costs if you moved them elsewhere?
“Considerations like these better capture the true costs of computation, taking it beyond the purely financial.”
The carbon footprint of an algorithm depends on two factors: energy needed to run it and the GHG emissions released per unit of energy needed. This depends on the computing resources needed (including how long it takes to run, the number of cores and memory used and how efficient the data centre is) and the data centre’s location and what powers it (e.g. nuclear, gas or coal). Switching from an average data centre to a more efficient one can reduce carbon footprint by a third.
Computations are rarely performed only once. Algorithms are run multiple times, sometimes hundreds, and many are for good reasons but frequently they are unnecessary. This also increases GHG emissions, sometimes substantially. Limiting the number of times an algorithm runs is perhaps the easiest way to reduce unnecessary carbon footprint.
Director of the NIHR Cambridge BRC Professor Miles Parkes said: “We are delighted to have supported such timely and vital research, which has created an open tool for quantifying the carbon footprint of virtually any computation.
“The NIHR is committed to combating climate change and helping the UK meet the goals set forth in the Climate Change Act 2008.
“NIHR both promotes green approaches to health research and actively designs new tools that expand opportunities for green research that can be translated into policy and action.
“This tool empowers researchers, patients and the public to understand and minimise the greenhouse gas emissions of both their work and leisure, whether using home laptops, a high-performance computing cluster or the cloud.”
Dr Inouye added: “We hope our study will help raise awareness of the environmental impacts of day-to-day computer use and the computations underlying data science as well as promote more sustainable practices within the data science community.
“We want to make green computing a cornerstone of carbon reduction within the NHS, ensuring that the artificial intelligence revolution supports both a healthier society and planet.”
- The research paper Green Algorithms: Quantifying the carbon footprint of computation by lead authors Loïc Lannelongue and Jason Grealey, and Michael Inouye, is available to view on the open access journal Advanced Science.
- Find out more about this research and the hidden costs of computations in Advanced Science’s feature article on Green Algorithms: Quantifying the carbon footprint of computation.
- The NIHR Carbon Reduction Guidelines have been published to help researchers explore how they can apply the principles of good carbon management and sensible study design to reduce carbon footprint.
- Three days of action to highlight urgency of climate redress took place from 20-22 April, culminating in Earth Day 2021 on 22 April.



