DARE UK Multi-party trusted research environment federation
Establishing infrastructure for secure analysis across different clinical-genomic datasets – frequently asked questions
Introducing the DARE UK Federated Genomics project
Many research institutions and data providers use computing environments known as ‘trusted research environments’ (TREs) as a way to safely analyse data. However, these individual TREs currently cannot easily be used in combination when researchers would like to make use of data that is held within different TREs. Moving data from one TRE into another across organisations is costly, time consuming and full of barriers when meeting different organisations’ governance policies. The ability to analyse datasets held across different organisations could be very valuable to supporting research.
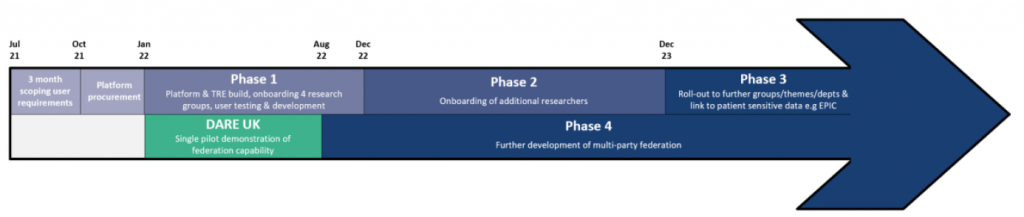
We have been funded by UK Research & Innovation as part of Phase 1 of the DARE UK (Data and Analytics Research Environments UK) programme, delivered in partnership with Health Data Research UK (HDR UK) and ADR UK (Administrative Data Research UK) to demonstrate a UK-first federation of genomic data by bridging the TRE of CYNAPSE, the new data infrastructure for the Cambridge Biomedical Campus, led by the University of Cambridge, with that of Genomics England.
We will do this by developing technologies that allow datasets held in these independent locations to be analysed simultaneously. The results of the separate analyses can then be combined without the original data ever having to move. The outcomes of this project will help establish new standards for federated TRE systems and unlock unprecedented possibilities for future collaborations with clinical-genomic data, potentially leading to new discoveries with long term public benefit.
Who is involved in the project and what are the timelines?
The project partners are the University of Cambridge, NIHR Cambridge Biomedical Research Centre, Genomics England, Eastern AHSN, Cambridge University Health Partners, and Lifebit. Phase 1 of the project started in January 2022, and it will run until the end of August 2022.
How does this fit into CYNAPSE?
The CYNAPSE programme will transform Cambridge’s computer infrastructure into a modern, future-proofed environment, supporting new approaches to clinical research across the Cambridge Biomedical Campus. One of the biggest components of the CYNAPSE programme will be to allow connectivity between datasets held within different research organisations TRE without the need to move datasets (known as ‘multi-party federation’).
The DARE UK Federated Genomic project, a stand-alone, separately funded project, will be a single pilot demonstration to test whether researchers can access and analyse data that is held in the two protected areas of CYNAPSE and Genomics England. The figure below shows where the DARE UK project fits into the CYNAPSE programme timeline.
What is federation of TREs?
Federation is the process of data being analysed using a piece of computer code (instructions to make the computer carry out a specific task). The code is designed to run the same analysis simultaneously in the separate TREs that hold the data researchers are interested in. The results are then brought together centrally in a separate TRE for final review, meaning the data never has to move.
What data is being used and where does it come from?
For the initial (proof of concept) project, both data sources are fully consented clinical-genomic data. Clinical-genomics is the study of clinical outcomes (measurable changes in health, function or quality of life that result from our care) with genomic data (study of the complete set of DNA in a person or other organism) The study of this data helps researchers understand the genetic basis for disease, resulting in more effective treatments.
The dataset from CYNAPSE is anonymous and already in the public domain and permission has been granted by Genomics England for the lead researcher of this project (Professor Serena Nik-Zainal) to run the analysis in question using the Genomics England dataset.
What are the benefits of TRE federation?
Data has the power to improve lives and has been fundamental to the UK’s response to the Covid-19 pandemic. Being able to support fast and efficient advanced analysis of data in an ethical and secure manner, whilst meeting the needs of researchers and data controllers, will support research at scale for the benefit of the public.
What governance procedures need to be addressed to allow for federation?
In the case of the DARE UK Federated Genomics project, the two datasets being used are from CYNAPSE, the data infrastructure for the Cambridge Biomedical Campus, and Genomics England. Both of the steps below would need to be completed before you would be able to federate the data between both organisations.
Requests for access to data from CYNAPSE would need to be approved by the Service Delivery Team who will check that the proposed access is safe and appropriate.
Genomics England datasets cannot be accessed without applying for access for a specific research project. Following approval by the Data Access Review Committee, and successful information governance training, researchers can access data within Genomics England’s secure research environment.
How do you ensure that data is not shared between the TREs?
The DARE UK system will form a virtual link between two TREs, with controls in place to ensure that data does not move between either location, only the combined results of the analysis. A preapproved computer code will allow the analysis to run in both TREs.
The main aim of the project is to show that it is possible to analyse the clinical genomic data safely and securely across these two locations using this method, leading to the development of a best practice model for future federated analytics.
Is there scope for this to be replicated with other organisations/datasets?
Yes, if successful, this initial prototype will serve as a blueprint, establishing the necessary best practices, frameworks, and open-source standards to inform how federated TREs can be used securely and transparently for large scale research.
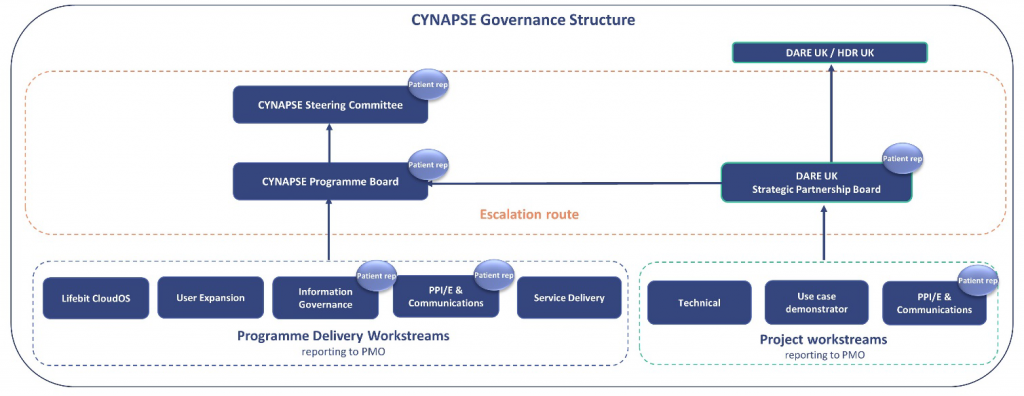
Is there a patient and public involvement and engagement (PPIE) element in the governance
structure or will there be in the future?
Yes, patient/public contributors from both NIHR Cambridge BRC and Genomics England have been involved in developing the governance and oversight framework of this project and we have patient representation within the project teams and attending project meetings.
Have people consented to the use of their data in this way?
The patient data in this project comes from individuals who have already consented to share their information for a particular research project. As this project will not be linking the two datasets but instead allowing for the results from the two to be analysed alongside each other (meaning the data does not move), the consent covers this process.


