Large-scale genetic study reveals new clues for the shared origins of irritable bowel syndrome and mental health disorders
An international study of more than 50,000 people with irritable bowel syndrome (IBS) has revealed that IBS symptoms may be caused by the same biological processes as conditions such as anxiety. The research highlights the close relationship between brain and gut health and paves the way for development of new treatments.
IBS is a common condition world-wide, affecting around 1 in 10 people and causing a wide range of symptoms including abdominal pain, bloating and bowel dysfunction that can significantly affect people’s lives. Diagnosis is usually made after considering other possible conditions (such as Crohn’s disease or bowel cancer), with clinical tests coming back ‘normal’. The condition often runs in families and is also more common among people who are prone to anxiety. The causes of IBS are not well understood, but an international team of researchers has now identified several genes that provide clues into the origins of IBS.
The research team, including more than 40 institutions and coordinated by scientists in UK and Spain, looked at genetic data from 40,548 people who suffer with IBS from the UK Biobank and 12,852 from the Bellygenes initiative (a world-wide study aiming to identify genes linked to IBS) and compared them to 433,201 people without IBS (controls), focusing on individuals of European ancestry. The findings were repeated with de-identified data from the genomics company 23andMe Inc., provided by customers who have consented to research, by comparing 205,252 people with IBS to 1,384,055 controls.
The results showed that overall, heritability of IBS (how much your genes influence the likelihood of developing a particular condition) is quite low, indicating the importance of environmental factors such as diet, stress and patterns of behaviour that may also be shared in the family environment.
However, 6 genetic differences (influencing the genes NCAM1, CADM2, PHF2/FAM120A, DOCK9, CKAP2/TPTE2P3 and BAG6) were more common in people with IBS than in controls. As IBS symptoms affect the gut and bowel, it would be expected that genes associated with increased risk of IBS would be expressed there – but this is not what the researchers found. Instead, most of the altered genes appear to have more clear-cut roles in the brain and possibly the nerves which supply the gut, rather than the gut itself.
Researchers also looked for overlap between susceptibility to IBS and other physical and mental health conditions. They found that the same genetic make-up that puts people at increased risk of IBS also increases the risk for common mood and anxiety disorders such as anxiety, depression, and neuroticism, as well as insomnia.
However, the researchers stress that this doesn’t mean that anxiety causes IBS symptoms or vice versa. Study co-senior investigator and consultant gastroenterologist Professor Miles Parkes (University of Cambridge) explained: “IBS is a common problem, and its symptoms are real and debilitating. Although IBS occurs more frequently in those who are prone to anxiety, we don’t believe that one causes the other – our study shows these conditions have shared genetic origins, with the affected genes possibly leading to physical changes in brain or nerve cells that in turn cause symptoms in the brain and symptoms in the gut.”

The study also found that people with both IBS and anxiety were more likely to have been treated frequently with antibiotics during childhood. The study authors hypothesise that repeated use of antibiotics during childhood might increase the risk of IBS (and perhaps anxiety) by altering the ‘normal’ gut flora (healthy bacteria that normally live in the gut) which in turn influence nerve cell development and mood.
Current treatments for IBS vary widely and include dietary changes, prescription medications targeting the gut or brain, or behavioural interventions. Lead author Chris Eijsbouts (University of Oxford) suggests that discovering genes which contribute to IBS may aid in the development of new treatments in the long term. He continues: “Even genetic changes that have only subtle effects on IBS can provide clues about pathways to target therapeutically. Unlike the individual genetic changes themselves, drugs targeting the pathways they tell us about may have a considerable impact on the condition, as we know from other disease areas.”
Co-senior investigator Dr Luke Jostins (University Oxford) commented: “We anticipate that future research will build on our discoveries, both by investigating the target genes identified and exploring the shared genetic risk across conditions to improve understanding of the disordered brain-gut interactions which characterise IBS.”
“IBS represents a remarkable challenge for genetic studies. These initial findings have been long awaited, and finally tell us this type of research is worth the struggle” concludes Ikerbasque Professor Mauro D’Amato from CIC bioGUNE, co-senior investigator and coordinator of the Bellygenes initiative.
This research received funding and support from National Institute for Health Research (NIHR) Biomedical Research Centres in Cambridge, Oxford, Nottingham and Manchester. Further funding and support was received from the Wellcome Trust, the Li Ka Shing Foundation and the Kennedy Trust for Rheumatology Research in the UK, and the Spanish Ministry of Economy and Competitiveness (Instituto Salud Carlos III), the Health Department of the Basque Government and the Swedish Research Council (Vetenskapsradet).
Hear more about this study from Professor Miles Parks.
Paper reference
Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders Nature Genetics
November 2021
New research shows how our brain uses nutritional state to control growth and age at puberty
Researchers have discovered how a receptor in the brain, called MC3R, detects the nutritional state of the body and regulates the timing of puberty and rate of growth in children and increases in lean muscle mass.
These findings, published in the journal Nature (on 3 November), may explain how humans have been growing taller and reaching sexual maturity earlier over the past century.
Researchers previously suggested it could be down to more reliable access to food for pregnant women and children, until now, precisely how the body senses its state of nutrition and turns that information into growth and sexual maturation had not been understood.
It was already known that signals reach the brain to indicate the body’s nutritional state, such as the hormones leptin, produced in adipose (fat) cells, and insulin, produced in response to increases in blood sugar levels. In a part of the brain called the hypothalamus, these hormones act on a small group of neurons that produce signals called melanocortins.
The melanocortins act on a variety of receptors, two of which are present in the brain. One of these, the melanocortin 4 receptor (MC4R) has previously been shown to regulate appetite and lack of MC4R results in obesity; however, the MC4R system does not control the effect of nutrition on growth and timing of puberty.
Now, a study, led by researchers from the MRC Metabolic Diseases Unit and the MRC Epidemiology Unit (both part of the Wellcome-MRC Institute of Metabolic Science) at the University of Cambridge, with collaborators from Queen Mary University of London, University of Bristol, University of Michigan and Vanderbilt University and supported by the NIHR Cambridge BRC, has discovered a role for the brain’s other melanocortin receptor, which is known as the melanocortin 3 receptor (MC3R).
They found that in response to nutritional signals the MC3R system controls the release of key hormones regulating growth and sexual maturation.
To show the link in humans, the scientists searched amongst the half a million volunteers in UK Biobank for people with naturally occurring genetic mutations that disrupt the function of the MC3R. They identified a few thousand people who carried various mutations in the gene for MC3R and found these people were on average shorter and went into puberty later than those with no mutation.
For example, they identified 812 women who had the same mutation in one of their two copies of the MC3R gene. This mutation only partly reduced the ability of the receptor to work. Despite this subtle effect, women who carried this were on average 4.7 months older at puberty than those without the mutation.
People with mutations that reduced the function of MC3R were also shorter and had lower amounts of lean tissue, such as muscle, but it had no influence on how much fat they carried.
To confirm these findings in children, they studied almost 6,000 participants from the Avon Longitudinal Study of Parents and Children (ALSPAC) and identified six children with mutations in MC3R. The six children were shorter and had lower lean mass and weight throughout childhood, showing that this effect starts very early in life.
All the people identified in these studies had a mutation in only one of the two copies of the gene. Finding mutations in both copies of the gene is vanishingly rare, but the researchers were able to identify an individual in the Genes and Health study with a very damaging mutation in both copies of the gene. This person was very short and went into puberty after the age of 20.
Professor Sir Stephen O’Rahilly, a senior author on the study and Director of the MRC Metabolic Diseases Unit at the University of Cambridge, and metabolism, endocrinology and bone theme lead at NIHR Cambridge BRC pictured right, said: “This discovery shows how the brain can sense nutrients and interpret this to make subconscious decisions that influence our growth and sexual development. Identifying the pathway in the brain whereby nutrition turns into growth and puberty explains a global phenomenon of increasing height and decreasing age at puberty that has puzzled scientists for a century.
“Our findings have immediate practical implications for the testing of children with serious delays in growth and pubertal development for mutations in the MC3R.

“This research may have wider implications beyond child development and reproductive health. Many chronic diseases are associated with the loss of lean mass, including muscle, with resultant frailty. This responds poorly to simple nutritional supplements such as protein-rich drinks. The finding that the activity of the MC3R pathway influences the amount of lean mass carried by a person suggests that future research should investigate if drugs that selectively activate the MC3R might help redirect calories into muscle and other lean tissues with the prospect of improving the physical functional of such patients.”

Professor John Perry, a senior author on the study from the MRC Epidemiology Unit at the University of Cambridge, pictured left, said: “This is such an exciting time for human genetics. By analysing the genetic sequences of large numbers of research participants, we can now understand fundamental biological processes that have remained elusive until now.”
The research was funded by the UK Medical Research Council, Wellcome and the National Institute for Health Research and supported by the NIHR Cambridge BRC.
Dr Rob Buckle, Chief Science Officer at the Medical Research Council, which was a funder of the research, said: “These findings have the potential to make a significant step forward in future management of disorders of growth and puberty, and improvements in the health of those suffering from frailty caused by chronic conditions. This study shows the value of long-term investment in both large UK population cohorts and multidisciplinary research to discover the underpinning causes of human health and disease.”
Scientists identify the cause of Alzheimer’s progression in the brain
Cambridge researchers have used human data to measure the speed of different processes that lead to Alzheimer’s disease and found that it develops in a very different way than previously thought. The results could help researchers to develop new treatments.
An international research team, led by the University of Cambridge and supported by the NIHR Cambridge BRC, found that Alzheimer’s disease starts in multiple, different regions of the brain, rather than from a single point which then spreads elsewhere. How quickly the disease kills cells in these regions determines how quickly the disease progresses overall.
The researchers used post-mortem brain samples from Alzheimer’s patients, as well as brain PET scans from living patients with a range of cognitive impairment, from mild impairment through to those with late-stage Alzheimer’s disease. The researchers used the samples and scans to track how a protein called tau (one of two key proteins thought to cause the condition) formed into clumps called ‘aggregates’.
In Alzheimer’s disease, tau and another protein called amyloid-beta, build up into tangles and plaques (clumps) – aggregates – causing brain cells to die and the brain to shrink. This results in memory loss, personality changes and difficulty carrying out daily tasks. For many years, the processes within the brain which result in Alzheimer’s disease have been described using terms like ‘cascade’ and ‘chain reaction’. It is a difficult disease to study, since it develops over decades, and a definitive diagnosis can only be given following examination of samples of brain tissue after death.
However, by combining five different datasets and applying them to the same mathematical model, the researchers observed that the speed at which aggregates multiply in individual regions of the brain determines the progression of Alzheimer’s disease, rather than aggregates spreading from one region to another.
“The thinking had been that Alzheimer’s develops in a way that’s similar to many cancers: the aggregates form in one region and then spread through the brain,” said Dr Georg Meisl from Cambridge’s Yusuf Hamied Department of Chemistry, the paper’s first author. “But instead, we found that when Alzheimer’s starts there are already aggregates in multiple regions of the brain, and so trying to stop the spread between regions will do little to slow the disease.”
The results, reported in the journal Science Advances, open up new ways of understanding the progress of Alzheimer’s and other neurodegenerative diseases, and new ways that future treatments might be developed.
This is the first time that human data has been used to understand the development of Alzheimer’s disease over time. It was made possible in part by approaches developed in Cambridge over the last decade, which allowed the modelling of protein aggregation and spread in the brain, as well as advances in PET scanning and improvements in the sensitivity of other brain measurements.
“This research shows the value of working with human data instead of imperfect animal models,” said co-senior author Professor Tuomas Knowles, also from the Department of Chemistry. “It’s exciting to see the progress in this field – fifteen years ago, the basic molecular mechanisms were determined for simple systems in a test tube by us and others; but now we’re able to study this process at the molecular level in real patients, which is an important step to one day developing treatments.”
The researchers found that tau aggregates multiply slower than expected – taking up to five years. “Neurons are surprisingly good at stopping aggregates from forming, but we need to find ways to make them even better if we’re going to develop an effective treatment,” said co-senior author Professor Sir David Klenerman, from the UK Dementia Research Institute at the University of Cambridge. “It’s fascinating how biology has evolved to stop the aggregation of proteins.
“The work allows us to determine the rate limiting molecular step in the development and spread of tau aggregate through the brain and hence should be targeted for an effective therapy for Alzheimer’s disease. In future it also might allow the effectiveness of a potential treatment to be measured by analysing the changes in PET signal over time using the model we have developed,” Professor Sir David Klenerman added.
The researchers say their research could be used to help the development of treatments for Alzheimer’s disease, which affects an estimated 44 million people worldwide, by targeting the most important processes that occur when humans develop the disease. It could also be applied to other neurodegenerative diseases, such as Parkinson’s disease.
“The key discovery is that stopping the replication of aggregates rather than their propagation is going to be more effective at the stages of the disease that we studied,” said Knowles.
The researchers are now planning to look earlier in the development of the disease, and extend the studies to other diseases such as Frontal temporal dementia, traumatic brain injury and progressive supranuclear palsy where tau aggregates are also formed during disease.
The study is a collaboration between researchers at the UK Dementia Research Institute, the University of Cambridge and Harvard Medical School. Funding is acknowledged from Sidney Sussex College Cambridge, the European Research Council, the Royal Society, JPB Foundation, the Rainwater Foundation, the NIH, and the NIHR Cambridge Biomedical Research Centre, which supports the Cambridge Brain Bank.
To read the full paper:
Georg Meisl et al.
‘In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease.’ Science Advances (2021).
DOI: 10.1126/sciadv.abh1448
Call for 10,000 volunteers in the UK to take part in international breast cancer study
NIHR Cambridge Biomedical Research Centre is supporting the UK arm of a large international study, to test out a more personalised way to screen for breast cancer and detect it sooner.
The study involves 6 European countries and is looking to recruit 85,000 volunteers aged between 50 and 70 who have never had breast cancer before – 10,000 volunteers are needed from the UK.
So far 3 NHS sites are involved in the trial, Cambridge University Hospitals NHS Foundation Trust (CUH), the Leeds Teaching Hospitals NHS Trust and Manchester University NHS Foundation Trust (MFT).
Professor Fiona Gilbert, professor of radiology at University of Cambridge, honorary consultant at CUH and NIHR Cambridge Biomedical Research Centre Imaging theme lead (pictured right), is leading the UK study.
She hopes the ‘MyPeBS’ trial, short for my personal breast screening, will see monitoring improved for all.
Professor Gilbert said: “This is an opportunity to take part in one of the largest studies so far into how we find early stage breast cancer. By taking a saliva sample and history from those selected on the trial, we can identify whether they are at higher or lower risk of developing breast cancer. Once we know this, we can tailor screening to their own personal needs.”

With almost 355,000 new cases diagnosed and 92,000 deaths each year in Europe, breast cancer is the most common and deadliest cancer in women, but it is most often curable if diagnosed early enough.
At the moment, all those aged 50 to 70 years are invited to participate in the NHS breast cancer screening programme by having a mammogram every three years.
However, not all are identical when it comes to breast cancer risk. Several factors including genetics, hormones, family history and breast density can put some at higher risk compared to others.
The MyPeBS study randomly assigns trial volunteers to follow either the standard NHS screening schedule or a personalised screening schedule according to their risk of breast cancer. The trial lasts for 4 years for all recruits.
Gareth Evans, professor in medical genetics and cancer epidemiology at Manchester University NHS Foundation Trust (MFT) and cancer prevention and early detection theme lead for the NIHR Manchester Biomedical Research Centre (BRC), leads the study in Manchester. He said: “We all carry tiny genetic variations, called single nucleotide polymorphisms (SNPs), and our unique combination of these can either raise or lower women’s risk of developing breast cancer, when combined with traditional risk factors like family and reproductive history, our health and lifestyle, and breast density.
“With this international trial we want to test how we can safely personalise breast screening depending on women’s individual risk, with women at lower risk safely screened less often, while those at high risk more frequently.”
The trial hopes to establish whether personalised risk-based screening could be more efficient and safer than the current, standard screening programmes, with fewer late-stage breast cancers diagnosed alongside fewer false positives and over-diagnoses. Researchers led by David French, professor of health psychology at the University of Manchester, will also be studying how women respond to changes in screening and any psychological impact this has, to help inform future screening services.
So far around 20,000 have joined the trial which started in summer. Around 1,000 recruits have joined the UK trial so far.
Dr Nisha Sharma, director of breast screening at Leeds Teaching Hospitals NHS Trust and the principle trial investigator in Leeds said: “We are excited to be part of this large scale and innovative trial. We would encourage women to contact the website if interested in taking part.”
The Cambridge site will be supported by the NIHR Cambridge Biomedical Research Centre.
Anyone interested in signing up can find information on the MyPeBs website www.mypebs.eu
The UK based charity Independent Cancer Patients’ Voice is a partner of the MyPeBS consortium who oversee the study.
ITV News reported on the story in October.
Adapted from CUH press release
Positive phase 3 results reported in trial for new Covid-19 vaccine
A clinical trial which took place at Cambridge University Hospitals for a new vaccine against Covid-19 has received positive Phase 3 results.
The trial has been taking place at 22 locations across the UK and was supported at the Cambridge site by the National Institute for Health Research (NIHR) Cambridge Clinical Research Facility and NIHR Cambridge Biomedical Research Centre.
Developed by the French specialty vaccine company Valneva and manufactured in Scotland, the vaccine is the only inactivated, adjuvanted Covid-19 vaccine in clinical development in Europe. This means, that like flu and polio vaccines, it contains dead versions of the virus that cannot cause disease.
The pivotal Phase 3, Cov-Compare trial recruited a total of 4012 participants aged 18 years and over, and 660 adolescents.
Results show that the vaccine was successful in producing high levels of neutralising antibodies against the disease.
Participants aged over 30 also reported fewer side effects within seven days of receiving the jab when compared to other vaccines, and no serious side effects were recorded.
Dr Effrossyni Gkrania-Klotsas, a consultant in infectious diseases at CUH and the local Principal Investigator for the trial said: “I am very pleased that we have been able to contribute to the development of another well-tolerated and effective vaccine option.
“We are very lucky to have a great NIHR Cambridge Clinical Research Facility that is extremely efficient in running vaccination trials, as well as a clinical team with extensive experience now from multiple vaccines. This is an investment towards future vaccine trials on the campus.”
Adam Finn, Professor of paediatrics at the University of Bristol, and Trial Chief Investigator added: “This is a much more traditional approach to vaccine manufacture than the vaccines so for developed in the UK, Europe and North America and these results suggest this vaccine candidate is on track to play and important role in overcoming the pandemic.”
Thomas Lingelbach, chief executive officer of Valneva said the results confirm the advantages which are often associated with inactivated whole virus vaccines.
He added: “We are committed to bringing our differentiated vaccine candidate to licensure as quickly as possible and continue to believe that we will be able to make an important contribution to the global fight against the COVID-19 pandemic. We are keen to propose an alternative vaccine solution for people who have not yet been vaccinated.”
Valneva hopes to initially get the jab approved for those aged between 18 and 55.
To find more visit the Valneva study website.
Professor Rebecca Fitzgerald awarded The Don Listwin award
Congratulations to NIHR Cambridge BRC researcher, Professor Rebecca Fitzgerald, who has received the Don Listwin Award for Outstanding Contribution to Cancer Early Detection.
The Don Listwin Award recognises a sustained contribution to, or singular achievement in, the cancer early detection field. The award, established in 2019, is named in honour of Don Listwin, founder and chairman of The Canary Foundation.
Rebecca Fitzgerald MD FMedSci, is internationally recognised for her exceptional research into the prevention and detection of oesophageal cancers. The Don Listwin Award honours the work she has done to develop, establish and grow the research needed to detect cancer early.
On receiving the award, Rebecca has paid credit to her team: “I am so honoured to be chosen for the 2021 Don Listwin Award and would like to thank all my fabulous team members who have believed in the early detection concept. We have still got a long way to go – but together we will make late diagnosis of cancer a thing of the past.”
Rebecca is known for the development of the Cytosponge technology, a sponge on a string that patients can swallow instead of undergoing an endoscopy. The Cytosponge collects cells from the oesophagus for use in a test which can flag the presence of TFF3-positive cells indicative of Barrett’s oesophagus, a precursor to oesophageal cancer. The cytosponge research was supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility.
Rebecca and her team have recently published work demonstrating that Cytosponge increases the identification of Barrett’s in individuals with frequent heart-burn symptoms by 10-fold compared to standard of care. The building of evidence for its clinical implementation for surveillance of high-risk individuals and in endoscopy sparing due to COVID-19 related pressures on health systems, continues to make a vital impact to patients’ lives.
The Scientific Programme Committee for this year’s Early Detection of Cancer Conference who are awarding the prize said: “Rebecca’s commitment to improving the wider research environment by enabling global research capabilities in oesophageal cancers and developing the research base in early detection through fostering collaborations are just a few of the reasons why we’re lucky to have her within the early detection community.” (Andrew Flewitt, Beverly Emerson and Alice Fan).
Rebecca is currently the Interim Director of the MRC Cancer Unit, Hutchison-MRC Research Centre, Professor of Cancer Prevention, and a Clinician Scientist leading research in the early detection of cancer for the University of Cambridge and the Cancer Research UK Alliance for Cancer Early Detection (ACED).
The award was received by Rebecca today at The Early Detection of Cancer Conference – an annual meeting which brings together global leaders in the field to discuss the early detection landscape and share ground-breaking research.
Could a cancer drug be key to helping patients recover from a heart attack?
A stage 2 trial is underway in Cambridge to investigate whether a cancer drug could improve the recovery of heart attack patients, by targeting the immune system.
The study led by researchers at Cambridge University Hospitals (CUH), the University of Cambridge and supported by the NIHR Cambridge Clinical Research Facility, has found that a low dose of the cancer drug, known as aldesleukin, injected into the skin of patients who have had an acute heart attack, increased the activation of immune cells shown to protect the heart.
In a previous study, the drug activates a rare white blood cell called innate type 2 lymphocyte (ILC2). ILC2 has previously been shown to decrease the harmful inflammation that promotes the build-up of fatty deposits in arteries.
By targeting the inflammation caused by the body’s immune response to a heart attack the researchers also hope to reduce a person’s chances of having a second heart attack. The Cambridge team are now conducting a Phase 2 clinical trial to test the drug.

Julian Hough (pictured right), had a heart attack in July despite having an active and healthy lifestyle. He’s on the road to recovery and wanted to take part in the trial. He said: “This trial is important because if this drug works, which is the results the doctors expect, it’s going to benefit so many people.
“For me personally it’s helped me get my confidence back. I can see the results of scans and blood tests at each stage of the trial as the weeks go by. I can see how things are progressing as I start to get back to normal life.”

Dr Rouchelle Sriranjan, honorary cardiology registrar and clinical research associate at CUH (pictured left), said: “Some patients who have heart attacks, have an imbalance in the cells of their immune system. These patients are at a higher risk of second heart attacks or strokes and have more damage done to the heart.
“The hope is a low dose of aldesleukin, will re-calibrate the imbalance in the immune system and promote healing of the heart muscle and lower inflammation in the blood vessels. We hope this drug will reduce a person’s chances of having a second heart attack.”
Dr Tian Zhao, BHF clinical lecturer in cardiovascular medicine at the University of Cambridge said: “Right now, there is no way to stop the immune system, which gets activated after a heart attack, from mistakenly damaging the heart.
“If our clinical trial shows that aldesleukin works by harnessing the ‘good cops’ of our immune system, we may have found a way to help the heart heal after a heart attack.”
Dr Joseph Cheriyan, CUH consultant clinical pharmacologist and chief investigator of the Phase 1b clinical trial added: “The findings represent very early positive signals but there’s still a long way to go. Work is currently ongoing in Phase 2 trials, which will hopefully lead to large scale Phase 3 trials in the next year or so.”
Professor Metin Avkiran, Associate Medical Director at the British Heart Foundation, who funded the study said: “Every five minutes someone is admitted to a UK hospital due to a heart attack. Thanks to research, heart attacks are now treatable, and seven out of ten people will survive. However, many heart attack survivors will still be left with damaged hearts.
“This research reveals a new approach that has the potential to both help heal hearts damaged by a heart attack and reduce the risk of a further heart attack.
“If clinical trials results confirm these early research findings, drugs that activate ILC2 could revolutionise heart attack treatment.”
The research is supported by NIHR Cambridge BRC and the Medical Research Council and featured on BBC Look East in October 2021.
Adapted from the British Heart Foundation press release
Supporting National Inclusion Week
National Inclusion Week runs between 27 September and 3 October 2021 with the aim to celebrate everyday inclusion in all its forms.
This is the 9th year that the campaign has been running and the theme for 2021 is ‘unity’. Here at the NIHR Cambridge BRC we are committed to being an inclusive research environment where we can share learning, best practices, successes and challenges with others in order to improve health research.
Below are some of our staff members who are supporting this year’s campaign.

“We want everyone to feel welcome on the Cambridge Biomedical Campus and for their contribution to research to be valued.
There are many fantastic initiatives in equality, diversity and inclusion that we can build on together, enhancing the richness of our collective experience and working with people of talent from all backgrounds.
We can and must all help to make this happen – and I thank everyone for their support in this.
Professor Miles Parkes,
Director NIHR Cambridge BRC
Consultant Gastroenterologist at Cambridge University Hospitals
“To achieve our Campus ambition of ‘doing great work in a great place to work’, we are continually seeking to build on our inclusive culture from diverse recruitment through to in-role support, training and career development opportunities for all.
Our collective participation is vital as I truly believe that everyone has a role to play in making this happen. “
Professor Nita Forouhi,
NIHR Cambridge BRC Diversity & Inclusion Lead and Director of Organisational Affairs at School of Clinical Medicine


“The NIHR BioResource has inclusivity as one of our core values.
We are united for inclusion to create an environment that supports all our staff to thrive, and in turn makes it simple and welcoming for everyone who wants to participate in research.”
Dr Nathalie Kingston,
Director of NIHR BioResource for Translational Research
“The NIHR Cambridge BRC is committed to inclusivity in research capacity building: nurses, midwives, allied health professionals, physicians, healthcare scientists from different backgrounds and from groups that may have had less involvement in research previously.
Inclusion benefits innovation and new ways of addressing problems and developing solutions in healthcare.”
Professor Christi Deaton,
NIHR Cambridge BRC Capacity Building Co-lead & Professor of Nursing


“I look forward to an NHS where research inclusivity is a routine agenda for all research, supported by adequate training and funding of essential staff with protected time, recognising the needs of minoritised groups.”
Dr Christopher Osuafor,
Honorary Specialty Registrar, Clinical Neurosciences
Further information:
Nurturing Inclusive Research at NIHR Cambridge Biomedical Research Centre
School of Clinical Medicine EDI events
Equality and Diversity Online Training – for University of Cambridge staff
Equality, diversity and inclusion at Cambridge University Hospitals
How is research helping with life-saving liver transplants?
As part of Organ Donation week, Professor Ludovic Vallier, theme lead for transplantation and regenerative medicine at the NIHR Cambridge BRC, talks about his team’s cutting edge liver research, why we need donated livers and the importance of collaborative working.
Professor Vallier explains that no donated livers are wasted even if they are not suitable for transplantation as researchers can use them to learn more about the liver and improve treatments to ultimately benefit patients.
Watch the short video below to find out about the latest research happening in Cambridge.
Journey of the Cytosponge
How does an idea for a new diagnostic test become a reality?
Follow the journey of the cytosponge or ’pill on a string’, a revolutionary new test to detect Barrett’s oesophagus, a condition that can lead to oesophageal (throat) cancer in a small number of people.
Created in Cambridge by Professor Rebecca Fitzgerald and supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility, the idea for the cytosponge began back in 2001. Twenty years later, this ground-breaking device could transform the way we test people for oesophageal cancer in a cheaper, less invasive way.
This short interactive document shows the crucial stages that research must go through to make sure any new medicines and devices are carefully tested to ensure they are safe for patients, before they are adopted into mainstream clinical practice.
Follow the journey of this research from ‘bench to bedside’, find out what the future holds for the cytosponge, and see how a research idea can become a reality, used today in the NHS.
Click on panel below and click through the interactive document. To exit, press escape (Esc) on your keyboard.
You can read an accessible format of the timeline here.
World first for AI and machine learning to treat Covid patients worldwide
In a groundbreaking study, Addenbrooke’s Hospital in Cambridge along with 20 other hospitals from across the world and healthcare technology leader, NVIDIA, have used artificial intelligence (AI) to predict Covid patients’ oxygen needs on a global scale.
The research was sparked by the pandemic and set out to build an AI tool to predict how much extra oxygen a Covid-19 patient may need in the first days of hospital care, using data from across four continents.
The technique, known as federated learning, used an algorithm to analyse chest x-rays and electronic health data from hospital patients with Covid symptoms.
To maintain strict patient confidentiality, the patient data was fully anonymised and an algorithm was sent to each hospital so no data was shared or left its location.
Once the algorithm had ‘learned’ from the data, the analysis was brought together to build an AI tool which could predict the oxygen needs of hospital Covid patients anywhere in the world.
Published today (15 September) in Nature Medicine, the study dubbed EXAM (for EMR CXR AI Model), is one of the largest, most diverse clinical federated learning studies to date.
To check the accuracy of EXAM, it was tested out in a number of hospitals across five continents, including Addenbrooke’s Hospital. The results showed it predicted the oxygen needed within 24 hours of a patient’s arrival in the emergency department, with a sensitivity of 95 per cent and a specificity of over 88 per cent.
“Federated learning has transformative power to bring AI innovation to the clinical workflow,” said Professor Fiona Gilbert (pictured right), NIHR Cambridge BRC Imaging theme lead, who led the study in Cambridge and is honorary consultant radiologist at Addenbrooke’s Hospital and chair of radiology at the University of Cambridge School of Clinical Medicine.
“Our continued work with EXAM demonstrates that these kinds of global collaborations are repeatable and more efficient, so that we can meet clinicians’ needs to tackle complex health challenges and future epidemics.”

First author on the study, Dr Ittai Dayan, from Mass General Bingham in the US where the EXAM algorithm was developed said: “Usually in AI development, when you create an algorithm on one hospital’s data, it doesn’t work well at any other hospital. By developing the EXAM model using federated learning and objective, multimodal data from different continents, we were able to build a generalizable model that can help frontline physicians worldwide.”
Bringing together collaborators across North and South America, Europe and Asia, the EXAM study took just two weeks of AI ‘learning’ to achieve high-quality predictions.
“Federated Learning allowed researchers to collaborate and set a new standard for what we can do globally, using the power of AI, ” said Dr. Mona G. Flores, Global Head for Medical AI at NVIDIA. “This will advance AI not just for healthcare but across all industries looking to build robust models without sacrificing privacy.”
The outcomes of around 10,000 Covid patients from across the world were analysed in the study, including 250 who came to Addenbrooke’s Hospital in the first wave of the pandemic in March/April 2020.
The research was supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre (BRC).
Work on the EXAM model has continued. Mass General Brigham and the NIHR Cambridge BRC are working with NVIDIA Inception startup Rhino Health, cofounded by Dr. Dayan, to run prospective studies using EXAM.
Professor Gilbert added: “Creating software to match the performance of our best radiologists is complex, but a truly transformative aspiration. The more we can securely integrate data from different sources using federated learning and collaboration, and have the space needed to innovate, the faster academics can make those transformative goals a reality.”
Researchers eye up new gene therapy trial that could reverse hereditary blindness
Restoration of sight from a rare genetic mutation may now become a reality thanks to a prestigious award from the National Institute for Health Research (NIHR) and Moorfields Eye Charity.
NIHR Cambridge Biomedical Research Centre (BRC) researcher and Addenbrooke’s Hospital Honorary Consultant Ophthalmologist, Dr Patrick Yu Wai Man, has received an NIHR Moorfields Eye Charity Advanced Fellowship award from the NIHR, in partnership with Moorfields Eye Charity, to undertake research into inherited optic neuropathies, which are genetic diseases that affect the optic nerve, causing progressive and irreversible blindness.
A major focus of this ambitious research programme will be on Leber Hereditary Optic Neuropathy (LHON), a disease caused by a gene mutation in the mitochondrial genome that triggers rapid loss of vision in mostly young adults.
Dr Yu Wai Man and his team will run a new clinical trial at Addenbrooke’s Hospital in Cambridge and Moorfields Eye Hospital in London, to better understand LHON and whether gene therapy can help restore sight in some people affected with this condition.
LHON and mitochondrial blindness
LHON is caused by genetic mutations in the mitochondrial genome – a unique piece of circular DNA that we inherit from our mother. As a result, healthy cells in the retina are lost, leading to optic nerve damage and severe loss of vision to the point where the person is registered blind. There are currently limited treated options for LHON.
LHON is classified as a rare disease, yet it is estimated to affect at least 1 in 30,000 people in the UK. Although women and children can be affected, the majority of cases occur in young men between the ages of 15 and 35. Most people are unaware that they carry a LHON mutation until symptoms begin to develop with blurred vision or if a family member is diagnosed with this condition.
The reasons why LHON predominantly affects men and why it starts so abruptly remains a mystery. “Your eyes are the greatest camera you’ll ever own and this disease is a devastating blow to people,” explains Dr Yu Wai Man, pictured right. “You can go from being fit and well and then suddenly, within weeks, your vision deteriorates rapidly, and you are told you can be registered as blind.

“We know that this disease is hereditary and affects mostly young men. But there are still so many unanswered questions about this condition and we need to find out more in order to identify suitable treatments. Our recent research published in Science Translational Medicine is promising, indicating that patients with LHON who have experienced loss of vision for up to one year can benefit from gene therapy. We now want to focus on people who have had this condition for more than one year and that is the basis of our trial.”
Launching the trial
The trial will use a form of gene therapy (where a healthy version of the gene is inserted into cells of the retina using a harmless virus) in patients who have lost their vision between one to five years (chronic LHON). It will be hosted across two sites at Addenbrooke’s Hospital and Moorfields Eye Hospital.
Dr Yu Wai Man said: “We know the disease quickly kills off healthy cells in the retina and we want to try and bring as many cells as possible from the brink to prevent further sight loss and potentially improve vision. In the pilot phase, we plan to recruit 30 patients who will be split into two equal groups. One group will receive the gene therapy injections in both eyes whereas the second group will not receive the treatment. The patients will then be closely monitored over two years to see if the treatment improves their eyesight. If we are successful, this breakthrough could be life-changing and it will also provide hope to people with visual impairment from other genetic diseases that affect the optic nerve.”
Monitoring disease and patient outcomes
The gene therapy trial is part of a wider research programme aimed at better understanding how the disease progresses over time (natural history) and what factors (biomarkers) predict the visual outcome in the inherited optic neuropathies. Dr Yu Wai Man and his team will make use of the latest technology available within the Cambridge Clinical Vision Lab, which has recently been established and supported by the NIHR Cambridge Clinical Research Facility and funded by the NIHR Cambridge BRC, as part of its advanced therapies initiative.
“The inherited optic neuropathies have a major impact on quality of life that goes beyond loss of vision,” Dr Yu Wai Man explains. “The Cambridge team will also develop a patient-reported outcome measure (PROM) that will allow the lived experiences of patients to be captured. This new tool will provide a more accurate picture of whether someone’s disease is progressing over time and it will be very helpful for future treatment trials when investigating whether a treatment is actually working.”
Setting sights on further research
Dr Yu Wai Man’s innovative research programme on the inherited optic neuropathies will be made possible thanks to funding from the NIHR in joint partnership with Moorfields Eye Charity. Dr Yu Wai Man said: “Without this level of commitment for rare diseases from the NIHR, we wouldn’t be able to conduct this kind of trial to try and save people’s sight.
“As a clinical academic, an NIHR Moorfields Eye Charity Advanced Fellowship award is a fantastic opportunity. This award will allow me to carry out research which I hope will make a difference to patients, and it will give me the opportunity to work with other leading researchers to further develop my skills in conducting translational research.
“My aim is to drive forward a programme of research in genetic conditions that affect the optic nerve. I’m hoping that our work will help pave the way for more gene therapy trials for other eye diseases – sooner rather than later.
“I’m very excited about what we’re going to learn and achieve in the next few years working closely with patient organisations. We may not be able to restore normal sight 100%, but if we could improve vision enough to have a positive impact on someone’s quality of life, that is the most important thing.”
More information about LHON and the clinical trial
- The most common cause of LHON is the 11778 mitochondrial DNA mutation
- This genetic defect affects the amount of energy being produced by mitochondria, which are the powerhouses that fuel cell activity in the human body
- Affected individuals with the 11778 mutation who have lost vision between one and five years will be eligible to take part in this new trial
Find out more information on the NIHR Fellowship Programme
Cambridge researchers win Croonian Medal and Lecture 2022
Congratulations to our Metabolism, Endocrinology and Bone theme lead, Professor Sir Stephen O’Rahilly and to Sadaf Farooqi, Professor of Metabolism and Medicine, who have been awarded the Croonian Medal and Lecture 2022 from the Royal Society.
The Croonian Medal and Lecture is the premier lecture in the biological sciences. The lectureship was created by William Croone FRS, one of the original Fellows of the Royal Society.
Professor Sir Stephen O’Rahilly and Professor Sadaf Farooqi were given this prestigious award for their seminal discoveries regarding the control of human body weight, resulting in novel diagnostics and therapies, which improve human health.
Professor Sir Stephen O’Rahilly said: “It is an enormous honour to have our work recognised in this way by the Royal Society, one of the world’s pre-eminent scientific societies. The list of previous Croonian Lecturers contains many legendary names and to find oneself on that list is as surprising as it is humbling.”
“I hope that the award of this highly prestigious prize to a pair of physician scientists will help inspire young doctors to bring their natural curiosity to bear on patients presenting to them with patterns of disease that seem to be atypical. The rigorous investigation of such patients can provide them with meaningful diagnoses and open up opportunities for novel approaches to their treatment, while also illuminating new areas of human biology.”
Professor Sadaf Farooqi said: “I am absolutely delighted that our work has been recognised by the Royal Society. To receive the award alongside my longstanding mentor Stephen O’Rahilly, who taught me how basic scientific principles can be applied to the study of clinical conditions, is a huge honour. This prestigious Award recognises our work as Clinician Scientists, the contributions of many team members past and present, our collaborators across the world and the patients and their families who have contributed to our research.”
Professor Sir Stephen O’Rahilly and Professor Sadaf Farooqi will be awarded their medal and prize at the Royal Society at a future date.
Link between amino acid and a range of common diseases could help predict personal risk
One of the first population-scale studies on how common genetic traits are influenced by variations in the DNA of mitochondria, the powerhouses of human cells, has been completed by scientists at the Wellcome Sanger Institute, the University of Cambridge, EMBL’s European Bioinformatics Institute (EMBL-EBI), and supported by the NIHR Cambridge BRC.
The team identified associations between mtDNA variants and an amino acid, N-formylmethionine (fMet), and effects of fMet on the risk of developing a range of common, late-onset illnesses.
The study, published (on 23 August 2021) in Nature Medicine found that higher fMet levels are associated with increased risk of a wide range of late-onset diseases and all-cause mortality, demonstrating fMet’s potential as a biomarker of ageing and disease risk, as well as the importance of research into mitochondrial DNA variants.
Mitochondria are organelles that are found in the cells of all complex organisms. They perform a number of vital biological functions, including the production of around 90 per cent of the energy that cells need to function. Mitochondria are unique in that they have their own genetic code, knowns as mitochondrial DNA (mtDNA), which is distinct from the DNA contained in the nucleus of every cell in an organism’s body1. mtDNA is passed on from mother to child.
Many common diseases are influenced by mitochondrial damage or disruption, including genetic diseases such as diabetes, heart disease and depression that are influenced by mutations in mtDNA.
Over time, the accumulation of mutations in mtDNA leads to distinct lineages in the population, known as haplogroups, which confer particular traits. Previous research has shown that haplogroup Uk, found in 10 per cent of the European population, is protective against diseases such as Parkinson’s Disease and ischaemic stroke (IS).
In this study, two large-scale datasets were analysed to look for associations between genetic variants in mtDNA and thousands of common molecular traits such as blood cell counts and plasma proteins, in order to understand the molecular mechanisms behind mtDNA associations with diseases.
In the INTERVAL dataset of up to 16,000 participants, the researchers identified significant associations between levels of fMet and mtDNA variants in haplogroups Uk and H32. The team then verified these associations using cellular models. When fMet levels were measured in a cohort of ischaemic stroke patients, they found lower fMet levels compared to those in a healthy control group.
The researchers then analysed data from EPIC-Norfolk, a study that tracked the health of participants over a 20-year period, to ask whether differences in fMet between individuals were associated with a wider-range of late onset diseases. In contrast to ischaemic stroke, higher fMet levels were associated with increased risk of illnesses such as kidney disease and heart failure.
Dr Cai Na, a first author of the study from the Wellcome Sanger Institute and EMBL’s European Bioinformatics Institute (EMBL-EBI), said: “We knew that the Uk haplogroup offered some protection against ischaemic stroke and Parkinson’s disease, and our findings suggests that variants in mitochondrial DNA that upregulate N-formylmethionine (fMet) may play a part in this protection. What was surprising is that these same variants are also associated with higher risk of other diseases. While further study of the molecular mechanisms at work is required, fMet does seem to be a promising biomarker that we could use to better predict an individual’s risk of developing a wide range of common diseases.”
Professor Patrick Chinnery, a senior author of the study from the University of Cambridge, said: “When we examined the molecular processes using human cellular models, we found that variants in Haplogroup Uk modulate protein synthesis and degradation in both the mitochondria and cytoplasm, and this affects cellular processes beyond mitochondrial bioenergetics. In the case of ischaemic stroke (IS), our findings suggest part of the protective effect of mtDNA haplogroup Uk may be attributed to reduced protein clearance mediated by fMet.”
Dr Aurora Gomez-Duran, a first author of the study from the University of Cambridge, said: “Our findings reveal the important part that mtDNA variants play in several pathologies, and that they play a deeper role in cellular homeostasis than we previously thought. Due to this, mtDNA should be carefully considered when investigating a diagnosis and delivering a treatment for age-associated diseases”
With only around 6,000 individuals on which to assess around 1,000 molecular traits in this study, other significant associations between mtDNA variants and genetic traits remain hidden for now. The next step will be to scale up this research in order to identify other traits associated with mtDNA variants.
Professor Nicole Soranzo, a senior author of the study from the Wellcome Sanger Institute, said: “Our study highlights the vast potential of large-scale, hypothesis-free research into mitochondrial DNA as a way of better understanding health and disease. N-formylmethionine (fMet) is a promising biomarker that could one day help us to monitor individual disease risk and plan pre-emptive interventions. This study has been a true collaborative effort with data from genome-wide association studies, cell lines and computational analysis combining to offer rich insights into human biology.”
New gene variants identified that cause hypertension in pregnant women
A new international study – jointly funded by an NIHR and Medical Research Council (MRC) partnership and supported by the NIHR Cambridge and Oxford BRC – has found a unique pair of gene variants that cause sudden onset high blood pressure in pregnant women.
Led by researchers from the Queen Mary University of London (QMUL) and St Bartholomew’s Hospital, the results have now been published in the journal Nature Genetics.
Hypertension (high blood pressure) affects 30% of adults. Most cases are caused by a combination of inherited and acquired factors that require long-term treatment to prevent the complications of stroke and heart attacks.
For one in ten people with hypertension, a specific cause can be found and removed. The most common cause is a tiny benign nodule in one of the adrenals. These are glands near the kidneys that produce steroid hormones.
The hormone aldosterone stimulates the kidneys to retain salt and hence increase blood pressure. As a result, the condition known as primary aldosteronism typically leads to a type of hypertension which is resistant to conventional drugs, and is linked to an increased risk of stroke and heart attacks compared to other patients with hypertension.
The research team have found a number of gene variants which cause the production of high levels of aldosterone from small adrenal nodules. Their latest study involves a discovery of a new type of primary aldosteronism caused by the coincidence of a unique pair of new variants which always occur together. The patients are predominantly women, who present with sudden onset of high blood pressure and low blood potassium in the early months of a pregnancy.
In partnership with Professor Christina Zennaro, Inserm Research Director at the Paris Cardiovascular Research Center, and colleagues in Paris, it emerged the new variants switch on a receptor molecule in the adrenal cells which recognises the pregnancy hormone Human Chorionic Gonadotropin (HCG), the same as is measured in routine pregnancy testing – and that the receptor molecule triggers a surge of aldosterone production.
Professor Morris Brown, Professor of Endocrine Hypertension at Queen Mary University of London said: “What was particularly satisfying is that recognition of the cause of hypertension in these women enabled them to complete a successful pregnancy, and that afterwards they were completely cured of hypertension by a procedure to remove the adrenal nodule, and were able to stop all their drugs.”
The study was jointly funded by the Efficacy and Mechanism Evaluation (EME) Programme, an MRC and NIHR partnership, and also received funding from the British Heart Foundation and Barts Charity.
Adapted from NIHR article
Artificial pancreas trialled for outpatients with type 2 diabetes for first time
An artificial pancreas could soon help people living with type 2 diabetes and who also require kidney dialysis.
Tests led by the University of Cambridge and Inselspital, University Hospital of Bern, Switzerland and supported by the NIHR Cambridge BRC, show that the device can help patients safely and effectively manage their blood sugar levels and reduce the risk of low blood sugar levels.
Diabetes is the most common cause of kidney failure, accounting for just under a third (30%) of cases. As the number of people living with type 2 diabetes increases, so too does the number of people requiring dialysis or a kidney transplant. Kidney failure increases the risk of hypoglycaemia and hyperglycaemia – abnormally low or high levels of blood sugar respectively – which in turn can cause complications from dizziness to falls and even to coma.
Managing diabetes in patients with kidney failure is challenging for both patients and healthcare professionals. Many aspects of their care are poorly understood, including targets for blood sugar levels and treatments. Most oral diabetes medications are not recommended for these patients, so insulin injections are the most commonly used diabetes therapy – though optimal insulin dosing regimens are difficult to establish.
A team at the University of Cambridge and Cambridge University Hospitals NHS Foundation Trust has previously developed an artificial pancreas with the aim of replacing insulin injections for patients living with type 1 diabetes. In research published in Nature Medicine, the team – working with researchers at Bern University Hospital and University of Bern, Switzerland – has shown that the device can be used to support patients living with both type 2 diabetes and kidney failure.
Unlike the artificial pancreas being used for type 1 diabetes, this version is a fully closed loop system – whereas patients with type 1 diabetes need to tell their artificial pancreas that they are about to eat to allow adjustment of insulin, for example, with this new version they can leave the device to function entirely automatically.
Dr Charlotte Boughton from the Wellcome Trust-MRC Institute of Metabolic Science at the University of Cambridge, who led the study, said: “Patients living with type 2 diabetes and kidney failure are a particularly vulnerable group and managing their condition – trying to prevent potentially dangerous highs or lows of blood sugar levels – can be a challenge. There’s a real unmet need for new approaches to help them manage their condition safely and effectively.”
The artificial pancreas is a small, portable medical device designed to carry out the function of a healthy pancreas in controlling blood glucose levels, using digital technology to automate insulin delivery. The system is worn externally on the body, and is made up of three functional components: a glucose sensor, a computer algorithm to calculate the insulin dose, and an insulin pump.
The team recruited 26 patients requiring dialysis between October 2019 and November 2020. Thirteen participants were randomised to receive the artificial pancreas first and 13 to receive standard insulin therapy first. The researchers compared how long patients spent in the target blood sugar range (5.6 to 10.0mmol/L) over a 20 day period as outpatients.
Patients using the artificial pancreas spent on average 53% of their time in the target range, compared to 38% when they used the control treatment. This equated to around 3.5 additional hours every day spent in the target range compared with the control therapy.
Mean blood sugar levels were lower with the artificial pancreas (10.1 vs. 11.6 mmol/L). The artificial pancreas reduced the amount of time patients spent with potentially dangerously low blood sugar levels, or ‘hypos’.
The efficacy of the artificial pancreas improved considerably over the study period as the algorithm adapted, and the time spent in the target blood sugar range increased from 36% on day one to over 60% by the twentieth day. This finding highlights the importance of using an adaptive algorithm, which can adjust in response to an individual’s changing insulin requirements over time.
When asked about their experiences of using the artificial pancreas, everyone who responded said they would recommend it to others. Nine out of ten (92%) reported that they spent less time managing their diabetes with the artificial pancreas than during the control period, and similar numbers (87%) were less worried about their blood sugar levels when using it.
Other benefits of the artificial pancreas reported by study participants included less need for finger-prick blood sugar checks, less time required to manage their diabetes resulting in more personal time and freedom, and improved peace of mind and reassurance. Downsides included discomfort wearing the insulin pump and carrying the smartphone.
Senior author Professor Roman Hovorka, also from the Wellcome Trust-MRC Institute of Metabolic Science, said: “Not only did the artificial pancreas increase the amount of time patients spent within the target range for the blood sugar levels, but it also gave the users peace of mind. They were able to spend less time having to focus on managing their condition and worrying about the blood sugar levels, and more time getting on with their lives.”
Dr Boughton added: “Now that we’ve shown the artificial pancreas works in one of the more difficult-to-treat groups of patients, we believe it could prove useful in the wider population of people living with type 2 diabetes.”
The team is currently trialling the artificial pancreas for outpatient use in people living with type 2 diabetes who do not need dialysis and exploring the system in complex medical situations such as perioperative care.
Dr Lia Bally, who co-led the study in Bern, said: “The artificial pancreas has the potential to become a key feature of integrated personalised care for people with complex medical needs.”
The research was supported by the NIHR Cambridge Biomedical Research Centre, The Novo Nordisk UK Research Foundation, Swiss Society for Endocrinology and Diabetes, and Swiss Diabetes Foundation and Swiss Kidney Foundation.
Paper reference:
Boughton, CK et al. Fully automated closed-loop glucose control compared with standard insulin therapy in adults with type 2 diabetes requiring dialysis: an open-label, randomised crossover trial.
Nature Medicine; 4 Aug 2021
How the public can influence health research – free information session
Patients and the public play a vital role in how research is carried out at the Cambridge Biomedical Campus. Their voice can shape new research priorities and improve how we conduct our studies.
Members of the public are invited to attend an online information session hosted by the NIHR Cambridge Biomedical Research Centre to hear about our research and how patients and members of the public can get involved.
Our Patient and Public Involvement Lead, Dr Amanda Stranks, will be joined by one of our Patient and Public Involvement panel members to discuss what we mean by public involvement in research and what it is like to be involved.
If you are interested in health research and what happens at the campus or you want to get involved, sign up for this free information session on 15 September.
‘Biological fingerprint’ in blood could help identify COVID patients with no symptoms
Cambridge researchers are able to identify people who have had COVID-19 even if they displayed no symptoms. They have developed a way to find markers in the blood several months after infection, even if the individual had only mild or showed no symptoms at all.
Most people who have COVID-19 may recover in a few weeks but there are some who will develop severe symptoms that can last for several months.
Current practice requires people to take a PCR test at the time of infection or an antibody test, looking at the immune cells, to reveal if someone may have previously had the virus but were asymptomatic. Now Cambridge researchers have discovered a biomarker – a biological fingerprint – in the blood of patients who previously had COVID-19.
This has led the team to receive £370,000 from the National Institute for Health Research (NIHR) to develop a COVID-19 diagnostic test that will complement existing antibody tests, as well as develop a test that could diagnose and monitor long Covid.
The research builds on a pilot project supported by the Addenbrooke’s Charitable Trust which has been recruiting patients from the Long COVID Clinic established in May 2020 at Addenbrooke’s Hospital.
Finding the biomarker
During the pilot, the team recruited 85 patients to the Cambridge-led NIHR COVID BioResource, which collects blood samples from patients when they are first diagnosed and then at follow-up intervals over several months.
In their initial findings, they identified a molecule known as a cytokine produced by T cells in response to infection. As with antibodies, this biomarker persists in the blood for a long time after infection.
Dr Mark Wills from the Department of Medicine at the University of Cambridge, who co-leads the team, said: “We need a reliable and objective way of saying whether someone has had COVID-19. Antibodies are one sign we look for, but not everyone makes a very strong response and this can wane over time and become undetectable.
“We’ve identified a cytokine that is also produced in response to infection by T cells and is likely to be detectable for several months – and potentially years – following infection. We believe this will help us develop a much more reliable diagnostic for those individuals who did not get a diagnosis at the time of infection.”
By following patients for up to 18 months post-infection, the team hopes to address several questions, including whether immunity wanes over time. This will be an important part of helping understand whether people who have been vaccinated will need to receive boosters to keep them protected.
As part of their pilot study, the team also identified a particular biomarker found in patients with long COVID. Their work suggests these patients produce a second type of cytokine, which persists in patients with long COVID compared to those that recover quickly and might be one of the drivers behind the many symptoms that patients experience. This might therefore prove to be useful for diagnosing long COVID.
Dr Nyarie Sithole, also from the Department of Medicine at the University of Cambridge, who co-leads the team and helps to manage long COVID patients, said: “Because we currently have no reliable way of diagnosing long COVID, the uncertainty can cause added stress to people who are experiencing potential symptoms. If we can say to them ‘yes, you have a biomarker and so you have long COVID’, we believe this will help allay some of their fears and anxieties.
“There is anecdotal evidence that patients see an improvement in symptoms of long COVID once they have been vaccinated – something that we have seen in a small number of patients in our clinic. Our study will allow us to see how this biomarker changes over a longer period of time in response to vaccination.”
At the moment, the team is using the tests for research purposes, but by increasing the size of their study cohort and carrying out further work, they hope to adapt and optimise the tests that can be scaled up and speeded up, able to be used by clinical diagnostic labs.
As well as developing a reliable test, the researchers hope their work will help provide an in-depth understanding of how the immune system responds to coronavirus infection – and why it triggers long COVID in some people.
Dr Sithole added: “One of the theories of what’s driving long COVID is that it’s a hyperactive immune response – in other words, the immune system switches on at the initial infection and for some reason never switches off or never goes back to the baseline. As we’ll be following our patients for many months post-infection, we hope to better understand whether this is indeed the case.”
In addition, having a reliable biomarker could help in the development of new treatments against COVID. Clinical trials require an objective measure of whether a drug is effective. Changes in – or the disappearance of – long-COVID-related cytokine biomarkers with corresponding symptom improvement in response to drug treatment would suggest that a treatment intervention is working.
Understanding COVID-19
An extensive programme of 15 new research studies, backed by government funding through the NIHR, will allow researchers across the UK to draw together their expertise from analysing long COVID among those suffering long-term effects and the health and care professionals supporting them. These groundbreaking studies aim to help those people affected return to their normal lives.
Health and Social Care Secretary, Sajid Javid, said: “Long COVID can have serious and debilitating long term effects for thousands of people across the UK, which can make daily life extremely challenging.
“This new research is absolutely essential to improve diagnosis and treatments and will be life-changing for those who are battling long-term symptoms of the virus.
“It will build on our existing support with over 80 long COVID assessment services open across England as part of a £100 million expansion of care for those suffering from the condition and over £50 million invested in research to better understand the lasting effects of this condition.”
Cambridge GPs launch ‘pill on a string’ cancer check
A Cambridge device to spot early signs of cancer is being trialled for the first time to GP patients in the UK.
The cytosponge or ‘pill on a string’ has been designed to help detect the early stages of oesophageal cancer.
Developed by the University of Cambridge and supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility, the device is now being trialled with GP patients in a mobile unit – the first of its kind.
Patients who are being treated for heartburn from Cambridgeshire GP practices will be some of the first in the UK to have the simple 10-minute test outside a hospital setting.
The hope is this new, quick and potentially life-saving device will detect the early signs of oesophageal cancer and treatment can begin a lot sooner.
What is the cytosponge?
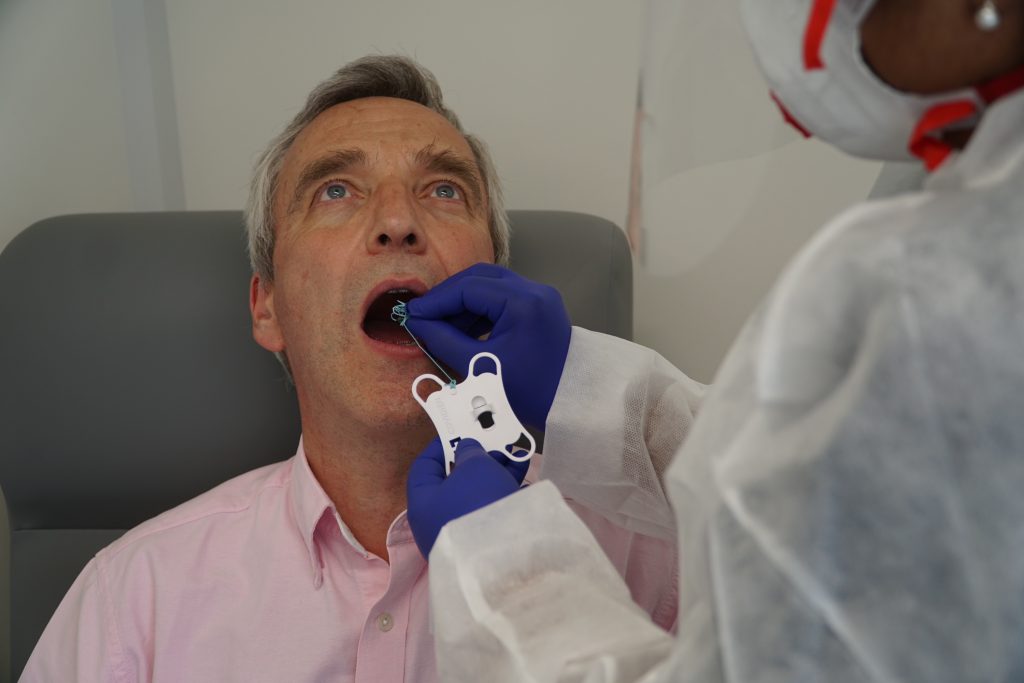
The cytosponge, developed by Professor Rebecca at the University of Cambridge, is a small pill with a thread attached that the patient swallows, which expands into a small sponge when it reaches the stomach.
It is quickly pulled back up the throat by a nurse, collecting cells from the oesophagus which will be sent for analysis.
The procedure takes around 10 minutes and can be performed in a GP surgery.
A recent Cancer Research UK funded medical trial picked up 10 times more cases of Barrett’s oesophagus, a pre-cancerous condition, than the GP’s usual first course of action.
The BEST3 trial involved more than 13,000 people and included research through GP practices which was facilitated by the Cambridgeshire and Peterborough Clinical Commissioning Group.
Now a cytosponge mobile unit known as the Heartburn Sponge Test unit (pictured top right) which has been funded and equipped jointly by Heartburn Cancer UK and Innovate UK funded Project DELTA will start seeing patients. After Cambridge it will move on to Essex and then Suffolk as the pilot aims at proving a wider benefit to the NHS.
Helping patients through research
One person who is thankful for the new device is Liz Chipchase from Cambridge.
Liz joined the BEST3 medical trial to help out. She’d been treated for acid reflux/heartburn for 40 years but didn’t expect any issues to show up. The results of the sponge test revealed she, in fact, had oesophageal cancer.
Fortunately – she feels ‘completely well thanks’ to the sponge test trial – it was found at an early stage and was treatable.
Liz was one of the lucky ones. Because oesophageal cancer is often found late, is more usually fatal. Only 17% of people diagnosed with it live for a further five years or more after diagnosis.
It is hoped the cytosponge could one day become a test used by GP surgeries throughout the country to identify potential issues for people who are on long-term heartburn medication, or when someone has had heartburn or indigestion for three weeks or more.
Genomics front and centre in blood matching
The international Blood transfusion Genomics Consortium launches its programme to expand cutting-edge genomics for more accurate blood typing, leading to improved transfusion therapy and better patient care
Blood Centres from around the world have united to form an international collaboration, supported by our partner the NIHR BioResource and a genomic array provider, Thermo Fisher Scientific, to deliver a project which promises to make future blood transfusion therapy more efficient, more accurate, standardised, and safer.
These organisations are collaborating in the Blood transfusion Genomics Consortium, which is coordinated by Cambridge University Hospitals and brings together medical experts in the treatment of anaemia and other blood cell disorders, blood transfusion, computer science, population statistics and genomics.
The main aim of the Consortium in this age of genomic medicine, is to validate for clinical use an affordable single DNA-based blood typing test, pioneered in the NIHR BioResource studies 07 and 89 to improve the match between donors of blood and the patients who receive the blood transfusion.
By working collaboratively between 10 countries across 4 continents, the adaptation of this new approach based on genomics, which was developed by the Consortium will for the first time set international standards for testing and for the use of blood transfusion therapy with the ultimate goal to improve the safety and efficacy of blood transfusion for the millions of patients we serve.
Each year, millions of donations of blood provide life-saving treatment to patients all over the world. Transfusion is essential to save the lives of patients with major bleeding (e.g. trauma), to treat patients whose blood production is impaired or reduced by chemotherapy (e.g. cancer), and to treat patients with inherited anemia who do not make sufficient haemoglobin of the healthy type (e.g. sickle cell disease).
Red blood cells have molecules on their surface called antigens which are inherited and differ between individuals. Current practice is to match blood for transfusion according to the patient’s major ABO and Rh blood groups, for which donor blood is currently tested. Antigens of the many other blood groups are not routinely matched unless the patient has a reaction, because the current tests are too expensive. Hence mismatches between the transfused donated cells and the cells of the patient’s not infrequently occur. For some, this can cause life-threatening reactions in which the transfused cells are rapidly destroyed by the immune system (antibodies) of the patient.
Dr Gleadall from Cambridge University Hospitals and NHS Blood and Transplant said: “It is amazing to see how the long-standing collaboration between the NIHR BioResource and Thermo Fisher Scientific has resulted in the development of a new method for testing the genes for blood groups, HLA and HPA types which is cost-effective and can be applied to both patient and donor blood samples”.
Blood types are determined by approximately 40 different genes. Consequently, DNA taken from a blood sample can be tested to determine the complete (more than ABO and Rh) blood type profile. The same test also allows us to rapidly type other tissue (HLA) and platelet (HPA) groups, which can help identify the best matched platelets to give to patients.
Dr Connie Westhoff from the New York Blood Center commented: “After decades of research we are now on the cusp of introducing a cutting-edge genomic technology into routine practice. This is an exciting development and will make it possible to find compatible blood for the many patients requiring regular transfusions, particularly those with sickle cell disease”.
Using samples from 8,000 blood donors, the results from the newly designed test have been compared with those of tests currently used by blood services and excellent agreement (99.9% concordance) was observed. Of note, almost halve of the small number (73) of differences were caused by record entry errors in the results for the currently used tests.
Professor Ellen van der Schoot from the Sanquin Blood Supply Foundation in the Netherlands said: “What is really exciting is that this new test results in a 10-fold increase in the number of available antigen types and is easy to implement allowing for its wide use. With data from patients with complex antibodies it has been shown that this increased typing information makes it more likely that a donation for safe transfusion can be identified. Indeed when the results of the new test obtained with the samples of 3,000 Dutch donors became available, Sanquin was able to provide life-saving transfusions to a young female patient for whom compatible blood could not be identified before”.
To bring this precision medicine test to the bedside, operational validation and regulatory acceptance is required. To deliver this the Consortium researchers have developed a well-defined study protocol with precise timelines for delivery. By using the samples and antigen typing data of almost 100,000 blood donors from the seven blood services collaborating in this study, it is anticipated that industry acceptance and regulatory approval can be completed over a 24-month period. Once validated, the test will be used to support clinical trials at hospitals supported by the seven blood services participating in the Consortium, in anticipation of introduction of the test platform internationally.
The Consortium participants hope to open up broader national and international access to a new generation of blood type analyses using genomic technologies to improve patient care around the globe. Organisations interested in joining are advised to visit the Consortium’s website at www.bgc.io to look whether they qualify as possible future Members of the Consortium.
