Children with cancer benefit from whole genome sequencing
More than 100 children with cancer from across the East of England have had their tumours tested by whole genome sequencing at Addenbrookes Hospital and supported by the NIHR Cambridge BRC, to help improve their diagnosis and treatment.
In cancer, Whole Genome Sequencing looks at the ‘whole genome’ or entire genomic (DNA) profile of a patient as well as the cancer.
For children with cancer, scientists look for differences, known as ‘variants’ or ‘mutations’, in the DNA from their tumour compared with their blood. This helps doctors and scientists give a far more detailed and personalised diagnosis, in some cases providing clues to the most effective treatments for each patient. Data from the first 36 children, who consented to the test as part of the national 100,000 Genome Project cohort, has now been published in the British Journal of Cancer.
The published findings, also shared at the 2021 National Cancer Research Institute (NCRI) Festival, described 23 different solid tumour types, and revealed several potentially important variants. In a number of cases, the information either refined or changed the children’s diagnosis, revealed new information about the children’s prognoses, showed hereditary causes, or revealed treatments that might not otherwise have been considered.
A further 65 patients across the region have had their whole genomes read since the test was made routinely available through the NHS Genomic Medicine Service at the start of 2021. Early review of the data and outcomes shows that these results continue to demonstrate the value of centralised WGS for children with cancer.
Aubrey, from Bedfordshire, was diagnosed with cancer in January 2021 when she was only 16 months old. However, as the actual type of Aubrey’s cancer was not certain from standard testing, her parents Anna and Paul agreed to a WGS test for Aubrey.

Anna, Aubrey’s mother said: “The test gave us a confirmed diagnosis for Aubrey after other tests had narrowed it down to one of two potential types of cancer. The result meant that the clinicians could be more confident as to the best treatment to use.
“Whist we still have a challenging journey with Aubrey’s diagnosis and treatment, we were relieved to know that she did not have cancer that was inherited, and hence we did not have to worry that it could affect our son or other members of the family as well.

Professor Matthew Murray, Honorary Consultant Paediatric Oncologist, Cambridge University Hospitals, pictured left, said: “Seeing 100 children with cancer benefit from WGS is a milestone. Overall, as a result of these tests, we’ve been able to confirm or refine the diagnosis for many of the children, identify and in some cases start new and beneficial treatment, and importantly in others have a clearer idea of the likely course of a patient’s cancer.”
Following referral, the NHS pathway allows patients and family to meet the clinical team at CUH to discuss the next steps.
Once consent to WGS had been obtained, a sample of tumour (usually taken from a previous procedure) is sent alongside a blood sample via the NHS East Genomic Laboratory Hub (NHS East GLH) to the company Illumina – located a few miles away from CUH. Samples are then sequenced at Illumina and results sent back for discussion at a meeting with the patient’s clinical team as well as expert scientists from the NHS East GLH to decide on best patient management.
This data has been released to coincide with the publication of the results from the patients enrolled in the 100,000 genome project in the British Journal of Cancer.
The research was supported by the NIHR Cambridge Biomedical Research Centre.
Adapted from CUH press release
Largest study of whole genome sequencing data reveals new clues to causes of cancer
DNA analysis of thousands of tumours from NHS patients has found a ‘treasure trove’ of clues about the causes of cancer, with genetic mutations providing a personal history of the damage and repair processes each patient has been through.
In the biggest study of its kind, a team of scientists led by Professor Serena Nik-Zainal from Cambridge University Hospitals (CUH) and University of Cambridge and supported by the NIHR Cambridge BRC, analysed the complete genetic make-up or whole-genome sequences (WGS) of more than 12,000 NHS cancer patients.
Because of the vast amount of data provided by whole genome sequencing, the researchers were able to detect patterns in the DNA of cancer, known as ‘mutational signatures’, that provide clues about whether a patient has had a past exposure to environmental causes of cancer such as smoking or UV light, or has internal, cellular malfunctions.
The team were also able to spot 58 new mutational signatures, suggesting that there are additional causes of cancer that we don’t yet fully understand.
This research was supported by Cancer Research UK and published on Friday 22nd April 2022 in the journal ‘Science’ here: http://www.science.org/doi/10.1126/science.abl9283
The genomic data were provided by the 100,000 Genomes Project an England-wide clinical research initiative to sequence 100,000 whole genomes from around 85,000 patients affected by rare disease or cancer.
Dr Andrea Degasperi, research associate at the University of Cambridge and first author said:
“WGS gives us a total picture of all the mutations that have contributed to each person’s cancer. With thousands of mutations per cancer, we have unprecedented power to look for commonalities and differences across NHS patients, and in doing so we uncovered 58 new mutational signatures and broadened our knowledge of cancer.”
Serena Nik-Zainal, a professor of genomic medicine and bioinformatics at the University of Cambridge, NIHR Cambridge BRC researcher and honorary consultant in clinical genetics at CUH said:
“The reason it is important to identify mutational signatures is because they are like fingerprints at a crime scene – they help to pinpoint cancer culprits. Some mutational signatures have clinical or treatment implications – they can highlight abnormalities that may be targeted with specific drugs or may indicate a potential ‘Achilles heel’ in individual cancers.”
“We were able to perform a forensic analysis of over 12,000 NHS cancer genomes thanks to the generous contribution of samples from patients and clinicians throughout England. We have also created FitMS, a computer-based tool to help scientists and clinicians identify old and new mutational signatures in cancer patients, to potentially inform cancer management more effectively.”
Michelle Mitchell, chief executive of Cancer Research UK, said: “This study shows how powerful whole genome sequencing tests can be in giving clues into how the cancer may have developed, how it will behave and what treatment options would work best. It is fantastic that insight gained through the NHS 100,000 Genomes Project can potentially be used within the NHS to improve the treatment and care for people with cancer.”
Professor Matt Brown, chief scientific officer of Genomics England said: “Mutational signatures are an example of using the full potential of WGS. We hope to use the mutational clues seen in this study and apply them back into our patient population, with the ultimate aim of improving diagnosis and management of cancer patients.”
Professor Dame Sue Hill, chief scientific officer for England and Senior Responsible Officer for Genomics in the NHS said: “The NHS contribution to the 100,000 Genomes Project was vital to this research and highlights how data can transform the care we deliver to patients, which is a cornerstone of the NHS Genomic Medicine Service.”
Adapted from CUH press release
Cambridge Clinical School successfully renews Silver Award for Athena Swan Charter
The University of Cambridge School Of Clinical Medicine, based at the Cambridge Biomedical Campus and part of the NIHR Cambridge Biomedical Research Centre partnership, has been awarded the Athena Swan Charter Silver Award for another five years.
The Athena Swan Charter is a framework used to support and transform gender equality within higher education and research. The Award celebrates good practice and recognises a commitment to making structural and cultural changes to advance gender equality.
The NIHR Cambridge BRC’s partnership with Cambridge University Hospitals and the University of Cambridge provides a strong platform to build on the key principles of the Athena Swan Charter for students and research staff. The renewal of the silver award demonstrates Cambridge’s ongoing commitment to gender equality, career progression and opportunities and making sure everyone is made to feel welcome on the biomedical campus.

Director of Organisational Affairs, Nita Forouhi, who led the School’s renewal application, pictured right, said: “The achievement of the Athena Swan Silver-level Award is pleasing indeed as it is a testament to the genuine commitment we give to equality, diversity and inclusion at the Clinical School.
“I am grateful to everyone who contributed to the huge effort towards the Silver Award renewal application and beyond that to everyone who is playing their part in improving the culture around our School to reach our ambition of ‘doing great work in a great place to work’. A special thanks is due to our Self-Assessment Team and its Chair, Graham Martin, to our EDI Governance Group members, to Caroline Newman and the HR EDI co-ordinators, to Fiona Karet the previous Director of Organisational Affairs and to our Departmental Equality Champions for their tremendous enthusiasm for the equality agenda.
“As much as it signifies our current efforts towards gender equality, this Award also means we have much work still to do. We look forward to doing this important work together.”

Professor Miles Parkes, Director of NIHR Cambridge Biomedical Research Centre, pictured left, said: “I am delighted to hear that our partner, the University of Cambridge School Of Clinical Medicine, has been awarded the Athena Swan Charter Silver Award.
“There are many fantastic initiatives in equality, diversity and inclusion that we can build on together, enhancing the richness of our collective experience and working with people of talent from all backgrounds.
“With our partners, Cambridge University Hospitals and University of Cambridge, the NIHR Cambridge BRC is building a research culture and environment where we can provide opportunities and career progression to all our research staff and make sure everyone is made to feel welcome on our campus.”
Further information on the Athena Swan Charter can be found here.
Brain charts map the rapid growth and slow decline of the human brain over our lifetime
An international team of researchers has created a series of brain charts spanning our entire lifespan – from a 15 week old fetus to 100 year old adult – that show how our brains expand rapidly in early life and slowly shrink as we age.
The charts are the result of a research project spanning six continents and bringing together possibly the largest ever MRI datasets ever aggregated – almost 125,000 brain scans from over a 100 different studies. Although not currently intended for clinical use, the team hopes the charts will become a routine clinical tool similar to how standardised paediatric growth charts are used.
Growth charts have been a cornerstone of paediatric healthcare for over 200 years and are used ubiquitously in clinics to help monitor the growth and development of children in comparison to their peers. A typical growth chart might plot age on the horizontal axis versus height on the vertical axis, but rather than being a single line, it will show a range that reflects the natural variability in height, weight or head circumference.
There are no analogous reference charts for measuring age-related changes in the human brain. The lack of tools for standardised assessment of brain development and aging is particularly relevant to the study of psychiatric disorders, where the differences between conditions and the heterogeneity within them demands instruments that can say something meaningful about a single individual in the way clinical reference charts can, and to conditions such as Alzheimer’s disease that cause degeneration of brain tissue and cognitive decline.
The study, published in Nature, is a major step towards filling this gap. Unlike paediatric growth charts, BrainChart – published on the open access site brainchart.io – covers the whole lifespan, from development in the womb through to old age, and aims to create a common language to describe the variability in brain development and maturation.
The incredible growing and shrinking brain
The brain charts have allowed the researchers to confirm – and in some cases, show for the first time – developmental milestones that have previously only been hypothesised, such as at what age the brain’s major tissue classes reach peak volume and when do specific regions of the brain reach maturity.
Dr Richard Bethlehem from the Department of Psychiatry at the University of Cambridge, one of the co-leads of the study, said: “One of the things we’ve been able to do, through a very concerted global effort, is to stitch together data across the whole life span. It’s allowed us to measure the very early, rapid changes that are happening in the brain, and the long, slow decline as we age.”
Among the key milestones observed by the team were:
- The volume of grey matter (brain cells) increases rapidly from mid-gestation onwards, peaking just before we are six years old. It then begins to decrease slowly.
- The volume of white matter (brain connections) also increased rapidly from mid-gestation through early childhood and peaks just before we are 29 years old.
- The decline in white matter volume begins to accelerate after 50 years.
- Grey matter volume in the subcortex (which controls bodily functions and basic behaviour) peaks in adolescence at 14-and-a-half years old.
Towards a clinically-useful tool
While the brain charts are already proving useful for research, in the long term, the team intend them to be used as a clinical tool. The datasets already have around 165 different diagnostic labels, meaning that researchers can see how the brain differs in conditions such as Alzheimer’s disease.
Alzheimer’s disease causes neurodegeneration and a loss of brain tissue, so people affected by the condition are likely to have reduced brain volume compared to their peers. In the same way that some healthy adults are taller than others, so there is variability in brain size – in other words, a slightly smaller brain does not necessarily indicate there is something wrong. However, as is apparent from the brain charts, while brain size decreases naturally with age, it does so much faster in Alzheimer’s patients.
Dr Bethlehem explained: “We’re still at an extremely early stage with our Brain Charts, showing that it is possible to create these tools by bringing together huge datasets. The charts are already beginning to provide interesting insights into brain development, and our ambition is that in future, as we integrate more datasets and refine the charts, they could eventually become part of routine clinical practice.
“You could imagine them being used to help evaluate patients screened for conditions such as Alzheimer’s, for example, allowing doctors to spot signs of neurodegeneration by comparing how rapidly a patient’s brain volume has changed compared to their peers.”
In addition, the team hope to make the brain charts more representative of the whole population, pointing to the need for more brain MRI data on previously under-represented socio-economic and ethnic groups.
A huge technical feat
Dr Jakob Seidlitz from the Lifespan Brain Institute at Children’s Hospital of Philadelphia and University of Pennsylvania, another of the co-leads of the study, said: “Creating these brain charts has involved multiple technical feats and a large team of collaborators. With brain imaging data, things are a bit more complicated than just taking out a measuring tape and measuring someone’s height, or head circumference. There were significant challenges to deal with, including logistic and administrative hurdles as well as the huge methodological variability we find between brain imaging datasets.”
The team used standardised neuroimaging software to extract data from MRI scans, beginning with simple properties such as the volume of grey matter or white matter, and then expanding their work to look at finer details, such as the thickness of the cortex or the volume of specific regions of the brain. They used a framework implemented by the World Health Organization for generating growth charts to build their brain charts.
Altogether, they estimate that they have used around 2 million hours of computing time, analysing close to a petabyte of data (a petabyte is equivalent to 1,000,000,000,000,000 bytes).
“This really wouldn’t have been possible without access to the High Performance Computing clusters at Cambridge,” said Dr Seidlitz. “But we still see this as a work in progress. It’s a first pass at establishing a standardised reference chart for neuroimaging. That’s why we’ve built the website and created a large network of collaborators. We expect to consistently update the charts and build on these models as new data becomes available.”
The team have created the tool with a reference framework to allow other researchers and clinicians to adjust their own datasets, making it possible to compare them against the BrainChart population.
Dr Bethlehem explained: “The NHS does millions of brain scans every year and in most of these cases, they are assessed by radiologists or neurologists relying on their extensive expertise to judge whether there is anything clinically relevant apparent on these scans. We hope that clinicians in future will be able to compare their data against ours and produce a more comprehensive report adding additional objective and quantitative observations to their assessment.
“This should effectively allow the neurologist to answer the question ‘this area looks atypical but atypical by how much?’. As the tool is standardised, it shouldn’t matter where you have your brain scan – you should still be able to compare it.”
Together with Dr Bethlehem and Dr Seidlitz, the work was led by Cambridge researchers Dr Simon White and Professor Ed Bullmore, and by Dr Aaron Alexander-Bloch at the University of Pennsylvania. It builds on a worldwide collective endeavour over the last few decades to measure human brain structure with MRI, in many different groups of people at many different ages. The team say it would not have been possible without open access to many high quality MRI datasets, and hope their results will contribute to further openness and sharing of data and analytics for brain imaging science.
The research was supported by the British Academy, the Autism Centre of Excellence, the Medical Research Council, National Institute for Health and Care Research (NIHR), the Wellcome Trust and the NIHR Cambridge Biomedical Research Centre.
Paper Reference
Bethlehem, RAI, Seidlitz J & White, SR et al. Brain charts for the human lifespan. Nature; 6 April 2022; DOI: 10.1038/s41586-022-04554-y
Cambridge recruits first patient to national inflammation study
A new trial taking place in UK to look at the best treatment for vasculitis has recruited its first patient– a child aged 10
A national trial led by chief investigator Professor David Jayne, NIHR Cambridge Biomedical Research Centre researcher and supported by the NIHR Cambridge Clinical Research Facility (CRF), is testing which biologic drugs – those that directly target the immune system – are most effective to treat vasculitis in all ages.
Vasculitis is a rare disease where inflammation occurs in the blood vessels. The immune system attacks the vessels, which become swollen, limiting or blocking blood supply to organs in the body.
Biologic drugs are licensed medicines and have been used to treat vasculitis and are expensive. Currently, there is not enough data to help clinicians choose which licenced drugs are most effective for patients, which is what the Cambridge team are investigating.
National trial begins
Vasculitis is potentially a life-threatening condition, yet the causes are unknown. Symptoms can range from fatigue and weight loss to numbness, pain and shortness of breath. Steroids are commonly used to help treat the disease, however, many people experience side effects or find no relief at all.
Cambridge researchers have now begun recruiting 140 adults and children around the UK with a rare vasculitis known as ANCA negative, who have not responded well to current treatments and cannot reduce their steroid dose to a safe level.
Patients will be randomised to either one of three biologic drugs or a placebo as well as answer questionnaires on their symptoms or if they have noticed any changes. They will then be monitored through follow-up appointments over a course of four to six months. The hope is to show which biologic medication is more effective.
Eliska, pictured top right with her mother, is the first child in the UK to take part in the trial at the NIHR Cambridge CRF at Addenbrooke’s Hospital. She said: “I’m hoping it will help me and also other children with vasculitis. I enjoy spending the time with the research nurses and Dr Armon.”
Kamila, Eliska’s mum, said: “I’m pleased that Eliska has a very special and experienced research team, and also good care here. I’m pleased they can help her and we hope she will be healthy soon.”

Dr Kate Armon, a paediatric rheumatologist, pictured left, who is overseeing the paediatric part of the trial in Cambridge said: “Vasculitis can be life-threatening and currently we don’t have enough information which medication is the most suitable. When we try to bring the steroid dose down for children and young people with vasculitis, the condition can flare, causing a fever, a rash and pain and stopping them from living a normal life.
“Our team agreed that the best option is to trial a biologic medication to see if it can control the disease better. On this trial, patients like Eliska will be randomised to a biologic drug as well as steroids and immune suppressive medication to control the illness, we will then monitor any changes to their condition.
“We hope to understand further how the biologic treatments affect patients like Eliska and find out which have better outcomes on the patient’s health and quality of life.
“I am delighted that Eliska has been able to participate in this important study to potentially treat her very rare disease. To have access to this trial in the NIHR Cambridge CRF has been perfect for the family and for the study.”
From physiotherapist to researcher: a new and exciting research career for Peter
‘Dr Hartley will see you now’ is a phrase that still sounds unfamiliar to physiotherapist, Peter Hartley. For the last 10 years, Peter has been busy working both clinically and academically to achieve his doctorate.
His extensive PhD research investigating muscle wastage in patients while in hospital, is just the beginning of an exciting future combining research with patient care. Here Peter shares his story and best tips for starting a career in research as an Allied Health Professional.
Going right back to the beginning, how did you start your career in research?

I completed my physiotherapy undergraduate degree at the University of East Anglia (UEA). Part of that was doing assignments and literature reviews, which was something I enjoyed, but in order to go into research, I was advised to have some clinical experience before pursuing it any further.
In 2011 I applied to do an NIHR Clinical Research Masters at the University of Hertfordshire. I hadn’t done any research activities in two years since my undergraduate studies, so it was nice to go back and do some academic work.
After that, I wasn’t sure what to do next. I was still seeing patients in the hospital, but I started questioning our current clinical practices – were there ways to prevent certain outcomes? Could I find new ways to improve patient care? I felt the best way to do that was via research.
I started working with researchers at the University of East Anglia to develop a doctoral application for the NIHR and focus my research in stroke patients. I spent a lot of time putting my application together, but unfortunately it was initially unsuccessful. However, the application experience gave me the skills and knowledge to know what to do for next time.
You undertook an NIHR Cambridge BRC and ACT fellowship, how did that help you?
At that point, I was moving my area of speciality and I began working in elderly medicine, so my research focus changed to match my clinical work. I started looking at how physiotherapy was beneficial to people on these wards and was this something I could investigate? I decided to apply for an NIHR Cambridge BRC and Addenbrooke’s Charitable Trust (ACT) fellowship and I was successful.
The BRC and ACT fellowship provided me with reserved time away from my clinical work to write research papers, carry out service evaluations, literature reviews, conduct PPI (patient and public involvement) work, attend conferences, work with the NIHR Research Design Service and think about my research proposal. It gave me the dedicated time and support I needed to put a strong doctoral application together.
The hardest part was deciding the actual research question. It was during my service evaluations and looking at the outcomes in older people who did not make a full functional recovery when leaving hospital that made me want to study this issue further. There was quite a bit of work conducting literature reviews to develop the argument to undertake research into this area and why it was important for patients. I found that as I was developing my proposal, my research question changed a lot.
As part of my application, I had to create a training programme and demonstrate how I would conduct the research which I enjoyed doing. Then I had to put together the protocols and methods that I would use to answer the research question. Finally, I had developed my idea and I was able to put my proposal forward and applied for my doctorate fellowship. I was fortunate that my application to the Dunhill Medical Trust was accepted.
Before my BRC and ACT fellowship, managing my clinical work and research applications was very challenging. The BRC and ACT fellowship gave me the protected time away from my clinical role to help me prepare and gather the evidence I needed for my doctorate application, without it I wouldn’t be where I am now.
What did your research study involve?
When I received my Dunhill Medical Trust fellowship I was working on my research full time. The first year I was studying, writing and doing a lot of reading, but in my second year I was recruiting patients to take part in the study and it was like being back in my clinical role.
With the support of the CRN (Clinical Research Network), I carried out an observational and an intervention study at Addenbrooke’s Hospital. The observational study had 70 participants who were patients on medical or geriatric wards and had unplanned admissions.
During the first week of their hospital admission I assessed patients’ strength every day and looked at their ability to move and walk, and measured how physically active they were during their hospital stay. Then 4-6 weeks after their discharge, I followed the patients up at home.
In the intervention study, 15 patients were randomised into two groups. I had planned to recruit more participants, but the trial then had to finish early because of Covid. Both groups were seen twice a day by a physiotherapy assistant. If they were in the intervention group, they were asked to perform simple strengthening exercises which included many repetitions of standing up and sitting down, which were progressed as the participants strength increased. Participants in the control group performed gentle stretches with the physiotherapy assistant.
What did you find out?
Before undertaking the research we knew that a large percentage of older adults have a reduced ability to manage their normal day to day activities when they leave hospital, compared with before they became unwell. We hypothesised that one of the main reasons for loss of muscle strength in hospital was due to inactivity.
We found on average, people lost around 10% of their muscle strength during their hospital stay. The more active someone was in the hospital, the more strength they retained. We also noticed the more frail someone was, the more their muscle strength reduced.
Were there any surprise findings?
The research did confirm our hypothesis, but what was interesting was that most of the patients had not recovered their muscle strength when I followed them up at home. There were also a few participants who were very mobile and independent before they were admitted to hospital and they were quite unwell in hospital, but we believed that they had made a good recovery and would resume their previous routines soon after they left hospital. However, we had not appreciated the impact that their hospital stay had had on their confidence, and when I visited them 4-6 weeks after they had been discharged home, they still had not left house in all that time. The impact on confidence was something I hadn’t given enough attention to before, but after that experience we decided to measure it with every patient.
With regards to the exercise study, we found patients supported the study, but after interviewing the participants, we identified some barriers to exercising in hospital. We discovered that far fewer of the exercise sessions were completed than we had anticipated. That was surprising because we assumed people who had signed up would be quite motivated to take part. It made us think about how we could produce a suitable exercise programme in the future and highlights the need to speak to patients or PPI groups to find out what will work best, and consider all the different factors that can affect a patient’s ability or motivation to exercise.
What happens next?
Our research has shown there is more work that needs to be conducted to improve muscle function in older hospital patients, but we’re hopeful this could be the beginning to making some changes in the physical activity of patients when they are admitted to hospital. It made us think about a couple of other things too. Could we work with patient groups and create an appropriate exercise programme for inpatients that is beneficial and acceptable to patients? Could we influence behaviour and encourage more activity and exercise on the ward in order to prevent loss of strength? Do we investigate other factors, such as building confidence for mobility when patients return home? And finally, investigating the economic factors of loss of muscle condition during hospital stays. Muscle wastage is quite important to our physical health – is it a bigger problem in the community after discharge? If we can consider exercise and reducing muscle loss on the ward, could it free up beds in a hospital because we are able to discharge patients who are more mobile? Would it prevent more people being discharged into care homes for short stays because their independence had increased? These are some of the areas we could explore in the future.
I’m now applying for various fellowships and grant awards for research funds to investigate these topics. I will also be returning to clinical practice and hopefully one of my applications is successful to let me continue my academic development.
What is it like having doctor in your title?
It’s still very odd having this title, I’m not used to the name yet and graduating was surreal because it was during COVID and I couldn’t attend the ceremony.
Looking back though, it has been a fantastic training opportunity and I have loved every minute. I would do it again in a heartbeat, it’s a hugely rewarding experience despite the challenges and rejections in the early days. The funding from the BRC and ACT fellowship was game-changing for me, it was so important. It gave me the time to put my application together and access the support I needed, I don’t think I would have been successful without it.
What advice would you give someone who was considering a research career?
Anyone who is thinking about going into research should do it. There is so much support out there if you’re willing to put the time in. I would say have a research question ready. Don’t worry if it changes a lot, it’s quite common as you’re developing your proposal.
If you don’t have a research question yet, think about the topic you would like to explore. Make use of the resources around you, speak to people, go to the library and do some reading to find out what research has already been done.
Contact your local Research Design Service, their advice is invaluable and they can help put you on the right path. We’re lucky in Cambridge, not only do we have good mentors but facilities such as the Clinical School library where I could work, join courses and seek advice. Look at funding, particularly the BRC and ACT fellowship that gives you the time to put your research application together.
Even though you may get knocked back a couple of times, don’t take it to heart. I found with rejection I learnt a lot about the application process and that is something that has been really useful and helped me with the next application.
Find yourself some good support and a mentor. Here in Cambridge I was supervised by Professor Christi Deaton, who is helping more nurses, midwives and AHP’s begin their research career. From my fellowship to now, Christi has been a great source of help with my applications and supported my doctoral studies.
My other doctoral supervisor, Professor Roman Romero-Ortuno, provided lots of support right from the beginning with my applications, publishing my first papers and helping with my doctoral and post-doctoral work. I very much doubt I’d be here without their help, and they have both been incredible.
“And be prepared!”
A lot can change along the way. When my research was accepted I thought I could just get everything organised and begin, but I found out that there can still be hurdles, even when the study has been setup. I had planned for a large number to take part, but I then found it was actually quite difficult to recruit people to the study for a number of reasons; patients discharge dates would change, people withdrawing from the study or people not wanting to take part because they had other things to consider while in hospital. It took me twice as long to get enough people enrolled in the trial than I expected. So, I had to go back and adjust my protocols that would fit in line with the study.
After making some changes the study took off. It was going well until the pandemic, where we had to pause the study and I went back into clinical practice for a while to support the hospital.
From my experience, it’s best to be prepared for the obstacles, think about every eventuality that could come your way, a lot can change on your research journey.
Looking back since then, it’s been a challenging road but it has been worth it, I wouldn’t change a thing and I’m looking forward to what the future holds.

Professor Christi Deaton, Professor of Nursing, who was Peter’s mentor said: “Peter is a shining example of what nurses, midwives and AHPs can achieve with support. His research is important for improving the outcomes of acutely hospitalised older adults and changing practice related to helping patients stay active in hospital.
“In addition to completing his PhD he has published several papers, been invited to speak internationally, led a Cochrane Collaboration Systematic Review, and is developing his post-doctoral research and collaborating with a company to improve activity monitoring. We will be hearing more from Dr. Hartley in future!
“Through our HEE (Health Education England) funding we have a website that has much information about clinical academic careers, funding, fellowships and the regional internship/bridging fellowship programme that I co-lead with Prof Tina Jerosch-Herald at UEA.”
Study suggests lithium may decrease risk of developing dementia
Researchers have identified a link which suggests that lithium could decrease the risk of developing dementia, which affects nearly one million people in the UK.
The researchers, from the University of Cambridge and supported by the NIHR Cambridge BRC, conducted a retrospective analysis of the health records of nearly 30,000 patients from Cambridgeshire and Peterborough NHS Foundation Trust. The patients were all over the age of 50 and accessed NHS mental health services between 2005 and 2019.
The analysis suggested that patients who received lithium were less likely to develop dementia than those who did not, although the overall number of patients who received lithium was small.
Their findings, reported in the journal PLoS Medicine, support the possibility that lithium could be a preventative treatment for dementia, and could be progressed to large randomised controlled trials.
Dementia is the leading cause of death in elderly Western populations, but no preventative treatments are currently available: more than 55 million people worldwide have dementia, with Alzheimer’s disease the most common form.
“The number of people with dementia continues to grow, which puts huge pressure on healthcare systems,” said Dr Shanquan Chen from Cambridge’s Department of Psychiatry, the paper’s first author. “It’s been estimated that delaying the onset of dementia by just five years could reduce its prevalence and economic impact by as much as 40 percent.”
Previous studies have proposed lithium as a potential treatment for those who have already been diagnosed with dementia or early cognitive impairment, but it is unclear whether it can delay or even prevent the development of dementia altogether, as these studies have been limited in size.
Lithium is a mood stabiliser usually prescribed for conditions such as bipolar affective disorder and depression. “Bipolar disorder and depression are considered to put people at increased risk of dementia, so we had to make sure to account for this in our analysis,” said Chen.
Chen and his colleagues analysed data from patients who accessed mental health services from Cambridgeshire and Peterborough NHS Foundation Trust between 2005 and 2019. Patients were all over 50 years of age, received at least a one-year follow-up appointment, and had not been previously diagnosed with either mild cognitive impairment or dementia.
Of the 29,618 patients in the study cohort, 548 patients had been treated with lithium and 29,070 had not. Their mean age was just under 74 years, and approximately 40% of patients were male.
For the group that had received lithium, 53, or 9.7%, were diagnosed with dementia. For the group that had not received lithium, 3,244, or 11.2%, were diagnosed with dementia.
After controlling for factors such as smoking, other medications, and other physical and mental illnesses, lithium use was associated with a lower risk of dementia, both for short and long-term users. However, since the overall number of patients receiving lithium was small and this was an observational study, larger clinical trials would be needed to establish lithium as a potential treatment for dementia.
Another limitation of the study was the number of patients who had been diagnosed with bipolar disorder, which is normally associated with an increased risk of dementia. “We expected to find that patients with bipolar disorder were more likely to develop dementia, since that is the most common reason to be prescribed lithium, but our analysis suggested the opposite,” said Chen. “It’s far too early to say for sure, but it’s possible that lithium might reduce the risk of dementia in people with bipolar disorder.”
This paper supports others which have suggested lithium might be helpful in dementia. Further experimental medicine and clinical studies are now needed to see if lithium really is helpful in these conditions.
The research was supported in part by the UK Medical Research Council and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre.
Paper reference:
Shanquan Chen et al. ‘Association between lithium use and the incidence of dementia and its subtypes: A retrospective cohort study.’
PLoS Medicine (2022). DOI: 10.1371/journal.pmed.1003941
Personalised blood test can detect persistent lung cancer
Patients who are at a higher risk of their lung cancer returning can be identified by a personalised blood test that is performed after treatment, according to researchers at the University of Cambridge.
Scientists at the Cancer Research UK Cambridge Institute and supported by the NIHR Cambridge BRC, used a personalised blood test for patients, which is a type of liquid biopsy that can pick up tiny fragments of DNA that are released into the blood as tumours grow. This DNA, called circulating tumour DNA (ctDNA), can reveal the state of the tumour, its location and potentially its weaknesses, which could be used to select the best treatments.
The results from the Lung Cancer Circulating Tumour DNA (LUCID-DNA) study, funded by Cancer Research UK, have been published today (Thursday 17th March) in the Annals of Oncology.
Many patients who are treated for early-stage non-small cell lung cancer can be cured with either surgery, radiotherapy or sometimes (chemo)radiotherapy.
After treatment, lung cancer patients are carefully followed up with tests including CT scans to find out if the treatment has removed the tumour, but scans won’t pick up tiny quantities of cancer cells known as minimal residual disease (MRD) which could still regrow into further tumours.
By finding signs that lung cancer cells might still be present and active after treatment, using methods such as liquid biopsy, doctors might be able make better choices about treating patients, aiming to improve the chances of survival for patients who are at higher risk while reducing side effects for patients who are at a lower risk group.
The LUCID-DNA study aimed to find out if circulating DNA can be detected in early stage lung cancers. It used a liquid biopsy, called RaDaR™, which analyses up to 48 different mutations that are unique to each patient’s tumour.
RaDaR™ was developed by Inivata, a biotech company co-founded by Dr Nitzan Rosenfeld, and is based on technologies developed initially by his lab at the Cancer Research UK Cambridge Institute.
Dr Nitzan Rosenfeld, group leader at the Cancer Research UK Cambridge Institute, Chief Scientific Officer of Inivata and co-lead author of the study, said:
“If cancer cells remain in the body after treatment a tumour can regrow. If that happens, it is a big setback for patients and the doctors treating them.
“Liquid biopsy can be used to detect tiny amounts of residual cancer after treatment, flagging those patients who have signs that their tumour may not have been eradicated completely with treatment. We’re hoping that this technology could help doctors decide when additional rounds of treatment are needed, and could save lives.”
To find out if liquid biopsy could find lung cancer patients with MRD, the LUCID-DNA study team enrolled 88 patients who were treated at Royal Papworth Hospital and Addenbrooke’s Hospital for early stage** non-small cell lung cancer (NSCLC). NSCLC accounts for over 85% of all lung cancer cases.
The research team extracted DNA from tumour samples provided by the patients and sequenced the DNA to find combinations of mutations unique to each patient’s lung cancer. Using this genetic “fingerprint”, Inivata created a blood test which was personalised to the patient’s tumour.
The liquid biopsies were then used to detect tumour DNA in blood samples collected before treatment, and for up to 9 months after treatment. The researchers found that patients who had tumour DNA present between two weeks and four months after treatment were much more likely to have their lung cancer come back or to die from it.
Professor Robert Rintoul, Professor of Thoracic Oncology at the University of Cambridge, Honorary Respiratory Physician at Royal Papworth Hospital and co-lead author of the study, commented:
“We need to study these liquid biopsies further to find the best ways to deploy them, but these results clearly show that they can potentially be an effective tool to help decide which patients need further treatment.
“Being able to offer patients personalised monitoring and treatment will ultimately save more lives and help us to beat cancer sooner.”
Lung cancer is the third most common type of cancer in the UK. Every year, around 48,500 people are diagnosed with lung cancer*** and every year around 35,100 people die from the disease in the UK****.
Aart Alders participated in the Lung Cancer Circulating Tumour DNA (LUCID-DNA) observational clinical study at Royal Papworth Hospital following his lung cancer surgery.
“I was first diagnosed with an early-stage lung cancer about five years ago and underwent a surgical operation to remove it,” he said. “Although some people need further treatment with chemotherapy, I have been very fortunate and my original lung cancer has not returned.
“I was very pleased to be able to help with the LUCID-DNA research study. By trying to develop a blood test to help doctors predict whether a lung cancer might come back or not, we will increase the chance of curing more people.”
Michelle Mitchell, Chief Executive of Cancer Research UK, said: “Lung cancer is one of the biggest killers in the UK. The earlier it is caught, the more likely it is to be treated successfully.
“Detecting signs of cancer before or after treatment without the need for invasive surgery has huge potential for both patients and doctors.
“I look forward to seeing more research that will develop liquid biopsy further, which will ultimately make it much easier for doctors to offer treatment that best matches the patient’s needs, increasing their chance of survival.”
Adapted from CRUK press release
Notes:
** The proportion of patients taking part in the study by stage of lung cancer at diagnosis was 48.9% at stage I, 28.4% at stage II and 22.7% at stage III.
*** Based on the annual average number of new cases of lung cancer in the UK between 2016 and 2018, available from https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer#heading-Zero (accessed 10/03/22).
**** Based on the annual average number of deaths from lung cancer in the UK between 2016 and 2018, available from https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer#heading-One (accessed 10/03/22).
Exploring how inflammation affects cognitive performance in people with depression
Research from Cambridgeshire and Peterborough NHS Foundation Trust (CPFT) and supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility, has found that inflammation may influence cognitive problems for people with depression, highlighting a new target to improve treatment.
The research team tested the cognitive performance of 80 people living with depression, and participants with evidence of inflammation had worse scores for processing speed and reaction time. Their results, reported in an international journal, will support further investigations to learn how inflammation is linked with cognitive dysfunction in depression, and how to effectively help people recover.
Lead author, CPFT consultant psychiatrist Dr Muzaffer Kaser said: “Cognitive problems are commonly experienced in depression, and new treatments targeting the immune system have potential to tackle these symptoms. Firstly, we need to better understand the link between inflammation and cognition in depression. In this exploratory study, depressed patients with high levels of inflammation markers showed poorer cognitive performance. Inflammation status may provide insights into how cognitive functions are affected in depression. The Insight Study will hopefully shed more light on how to best treat cognitive dysfunction in depression.”
The full article ‘Neurocognitive Performance in Depressed Patients with low-grade inflammation and somatic symptoms was published in Brain, Behavior, & Immunity – Health and is available online to read.
CPFT is also working with the Inflammation and Psychiatry Research Group on the Insight Study to further explore the links between inflammation and depression and find new treatments. Anyone aged 20-55 with depression and currently taking an antidepressant is invited to take part. If you would like to join, or you know someone who might be able to, please contact CPFT’s specialist Windsor Research Unit team running the study: email wru@cpft.nhs.uk or call 01223 219531.
The Insight Study lead Professor Golam Khandaker said: “The aim of this research is to generate knowledge about new causes of depression in order to develop better treatments. There is still time to take part, so please get in touch!”
The research on neurocognitive performance in depression was supported by CPFT, the University of Cambridge Department of Psychiatry and the National Institute for Health Research (NIHR), specifically NIHR Cambridge Clinical Research Facility, and NIHR Cambridge Biomedical Research Centre. The researchers received funding from a Wellcome Trust fellowship and NIHR Clinical Lectureship, the BMA Foundation, MQ: Transforming Mental Health, and the Medical Research Council UK.
Breaking the Bias for International Women’s day
International Women’s Day is celebrated globally on 8 March every year. Its focus is on highlighting women, calling for equal opportunities and removing discrimination. The theme for 2022 is ‘Breaking the Bias’.
We spoke to some of the women working in research in Cambridge, who told us what ‘Breaking the Bias’ means to them. Find out more about their role and why they believe International Women’s Day is important.

Vivien Mendoza – Matron, NIHR Cambridge Clinical Research Facility
“It means getting involved in a discussion and decision making, taking into consideration the skills of the staff involved: get others’ opinions and work as a team – and make use of each other’s strengths and consider each other’s weaknesses.”
Read more about Vivien’s role at the CRF and how she went from a neurosciences ward to working in research.
Tracy Cripps, Senior Research and Development Project lead
Watch the full video from Tracy on why International Women’s Day is important and on facing and overcoming bias in the workplace.

Debbie, Governance and Ethics Co-ordinator, NIHR BioResource
“It’s important to continue the push for women to be empowered to understand they can do it. Woman can be incredibly hard on themselves and that shows in a lack of confidence. International Women’s Day is a great platform to show women that actually yes you can do it, we as women can achieve and inspire life changing things.”

Jo Piper, Manager, NIHR Cambridge Clinical Research Facility
“International Women’s Day is a celebration of amazing women throughout history, championing their achievements. I think this theme is to ensure that we all challenge any ‘biased’ behaviour- everyone should be treated the same free of bias, stereotype or discrimination. Science / research should be open to all who want to work in this field and should not discriminate against anyone.”
Find out what this year’s theme ‘Break the Bias’ means to Jo.
Professor Tamsin Ford, Professor of Child and Adolescent Psychiatry
Watch Professor Ford explain the importance of visible role models for International Women’s Day.

Professor Nita Forouhi, Programme Leader and Honorary Consultant
“There are many conscious and unconscious biases related to gender and ‘Break the Bias’ is a call to recognise and acknowledge that fact and take action to reduce such biases.”

Rose Eichenberger, Governance and Ethics Manager,
NIHR BioResource
“‘Break the Bias’ means a lot to me – and it’s great to see the younger generation of female scientists being confident and present at the forefront of science.”
Find out about Rose’s role and how she was determined to work in research.
Anne Elmer, Matron, NIHR Cambridge Clinical Research Facility
Anne meets research teams everyday, she has noticed interesting choices when it comes to choosing a career in research.
Watch this short video, which also features a response from NIHR Cambridge Biomedical Centre Director Professor Miles Parkes on how we are achieving near parity in senior research roles held by women. See also Prof Parkes’s comment below.

Professor Miles Parkes, Director of NIHR Cambridge BRC, comments on the importance of International Women’s day and how more women are working in senior roles in Cambridge.
“International Women’s Day is an opportunity to highlight disadvantages that women may face in their careers and professional lives, as well as celebrate the enormous contribution women make to research.
“Across our NIHR Cambridge BRC organisation, many senior roles or positions of influence are held by women such the leads for our Office for Translational Research, for our Cambridge Clinical Trials Unit and our legal team.
“Since our recent organisational refresh, almost 50% of our theme leads and members of the senior executive committee are women. These women are at the top of their fields; they are world-class scientists and are the right people for the job.
“We’re building a research culture and environment such that everyone (not just based on gender, but all aspects of life and diversity) is made to feel welcome on our campus.”
Cambridge Professors receive NIHR Senior Investigator awards
Congratulations to Professor Emanuele Di Angelantonio, Professor of Clinical Epidemiology and Donor Health and Professor Tamsin Ford, Professor of Child and Adolescent Psychiatry, who have both been newly appointed as Senior Investigators for the NIHR.
Each year the NIHR invites applications into a round of open competition to decide who it awards the prestigious research roles to, based on outstanding contributions to research.
This year, 30 researchers have been newly-appointed to the NIHR Senior Investigator role, two of these have been awarded to NIHR Cambridge BRC researchers.
Professor Emanuele Di Angelantonio said: “I am honoured and proud to receive this recognition from the NIHR. The NIHR has been vital to my research into the fields of cardiovascular disease and blood donation, and I am excited to be able to contribute significantly as senior leader and ambassador.”
Professor Tamsin Ford said: “I am delighted and honoured to receive this award and looking forward to joining the NIHR academy.”
As part of the announcement, Professor Ian Wilkinson, Professor of Cardiology and Cardiovascular Disease, Professor Rebecca Fitzgerald, Professor of Gastroenterology and Hepatology, and Professor Martin White, Professor of Public Health are reappointed as Senior Investigators.
Professor Edward Bullmore, Mental Health and Psychiatry has been awarded Emeritus Senior Investigator.
Find out the full details of the 2022 award winners.
Cancer drug gives hope in treating heart attacks
Cambridge researchers have found a drug used to treat cancer could help with heart attack recovery.
Funded by the British Heart Foundation, NIHR Cambridge BRC and supported by the NIHR Cambridge Clinical Research Facility, researchers from Cambridge University Hospitals and the University of Cambridge found that a low dose of the cancer drug, aldesleukin, could harness the power of the immune system to improve recovery after a heart attack. Published in the New England Journal of Medicine – Evidence, researchers have said it has the potential to become the first treatment of its kind available for patients.
A heart attack is where the supply of blood is blocked to the heart, it can be life-threatening, requiring urgent treatment as it could potentially cause serious damage to the heart.
When a heart attack occurs it triggers the body’s immune cells to rush to the damaged heart and surrounding blood vessels. However, instead of having a healing effect, this can cause further harm, increasing the risk of future heart attack, stroke, and heart failure. Right now, there are no treatments available to counter this damaging immune response.
Cambridge researchers launched a 2a clinical trial at NIHR Cambridge Clinical Research Facility, where they tested aldesleukin, a drug used to treat cancer, to see if it could help the immune system and heart attack recovery.
High doses of aldesleukin stimulate the immune system to attack cancer cells. Researchers investigated whether using doses a thousand times lower than those used in cancer treatment could selectively target and boost anti-inflammatory cells in patients’ immune systems.
They found that low doses could improve recovery after a heart attack by stopping the harmful feedback loop. Researchers will now carry out larger clinical trials, in the hope it could be used to treat patients within the next 5 years.
Repurposing cancer medication
The study involved 16 patients admitted to hospital with a heart attack who were given one of two doses of aldesleukin or a placebo. The drug was injected under the skin in their abdomen once a day for five days, and they were then followed up again a week after they’d received the final dose of the drug.
Researchers found patients that received aldesleukin had a significantly greater increase in the number of regulatory T cells, a type of white blood cell that calms inflammation, a week after their last dose of aldesleukin compared to those who received a placebo.
Further analysis revealed that not only were the numbers of regulatory T cells increasing, but the cells themselves had features that suggested that they were also becoming more anti-inflammatory.
Low doses of aldesleukin also decreased the levels of other types of immune cells that can have damaging effects on inflammation and recovery after a heart attack. The team thinks this is another way that the drug could promote healing.
Researchers were encouraged with the results and are currently halfway through a larger clinical trial to investigate whether low doses of aldesleukin after a heart attack can reduce inflammation in patients’ blood vessels, which could potentially provide even more treatment to patients.
Dr Tian Zhao, British Heart Foundation Clinical Lecturer in Cardiovascular Medicine at the University of Cambridge, said: “It’s only in the past decade that we’ve begun to understand the considerable role that the immune system plays in heart attack recovery.
“In this study we’ve shown, for the first time, that low doses of aldesleukin given to heart attack patients can enhance the number of anti-inflammatory cells in the immune system. Previous research has suggested that this can reduce inflammation in blood vessels and improve heart healing.
“Our ongoing study will give us the first signs of whether this is having clinical benefits for patients. We hope these results will bring us one step closer to the first treatment that can stop the damaging immune response that follows a heart attack.”
Professor James Leiper, Associate Medical Director at the British Heart Foundation, said: “In the UK one person is admitted to hospital with a heart attack every five minutes. Thankfully, more people than ever are surviving heart attacks, but some will be left with long-term health problems such as heart failure. We urgently need new treatments that can help people to make a better recovery after a heart attack and reduce their risk of future ill health.
“Treatments that can unlock the anti-inflammatory power of the immune system have the potential to become a new treatment option for heart attack patients. This research is an important step towards making this type of treatment a reality.”
This research was also funded by the Medical Research Council.
Adapted from BHF press release
1,000 Covid patients sign up to UK drug trial
An Addenbrooke’s led study supported by the NIHR Cambridge BRC and NIHR Cambridge Clinical Research Facility, has enrolled a thousand Covid patients to find new treatments for the long term consequences of the disease and reduce the number who die months after leaving hospital.
The clinical trial – named HEAL-COVID – also aims to cut the number patients being readmitted to hospital with complications as a result of having Covid.
Data from the Office for National Statistics (ONS) suggests that around 1 in 10 patients with Covid-19 die in the six months after they leave hospital and more than 1 in 5 develop new or worsened symptoms during the first three months after discharge.
HEAL-COVID stands for Helping to Alleviate the Longer-term consequences of Covid-19 and is funded by the National Institute for Health Research (NIHR) and the NIHR Cambridge Biomedical Research Centre, and supported by the NIHR Cambridge Clinical Research Facility.
It is a clinical trial testing a number of safe, existing drugs to see if they can improve the longer-term outcomes for patients who have been hospitalised due to COVID-19.
Professor Charlotte Summers, pictured top right, who is leading the trial added: “Having survived the trauma of being hospitalised with Covid-19, far too many patients find themselves being readmitted with new or long term complications.
“Having launched this trial in March 2021, we still have thousands of people being treated in hospital due to Covid-19, and we will continue to offer them the opportunity to participate in HEAL-COVID so we can find treatments to improve their longer-term clinical outcomes.”

Dr Mark Toshner, honorary consultant at CUH, pictured left, added: “We are delighted that patients throughout the UK have access to this important study which remains the world’s largest post-hospital trial looking to reduce the longer term consequences of Covid-19.”
The trial is being led by Cambridge University Hospitals NHS Foundation Trust (CUH) and University of Cambridge, in collaboration with Liverpool Clinical Trials Centre (University of Liverpool) and Aparito Limited.
HEAL-COVID enrols patients when they are discharged from hospital, following their first admission for Covid-19.
They are randomised into 3 arms of the trial, receiving either the drug apixaban or atorvastatin or the usual care – and their progress tracked.
It’s hoped a fourth arm of the trial will be introduced soon, with a third drug being added on the recommendation of the UK COVID Therapeutic Advisory Panel (UK-CTAP).
Adapted from Cambridge University Hospitals article.
Artificial pancreas proves ‘life-changing’ for very young children with type 1 diabetes and their families
An artificial pancreas developed by a team of Cambridge researchers and supported by the NIHR Cambridge Biomedical Research Centre is helping protect very young children with type 1 diabetes at a particularly vulnerable time of their lives. A study published today (20 January) found that it is both safe to use and more effective at managing their blood sugar levels than current technology.
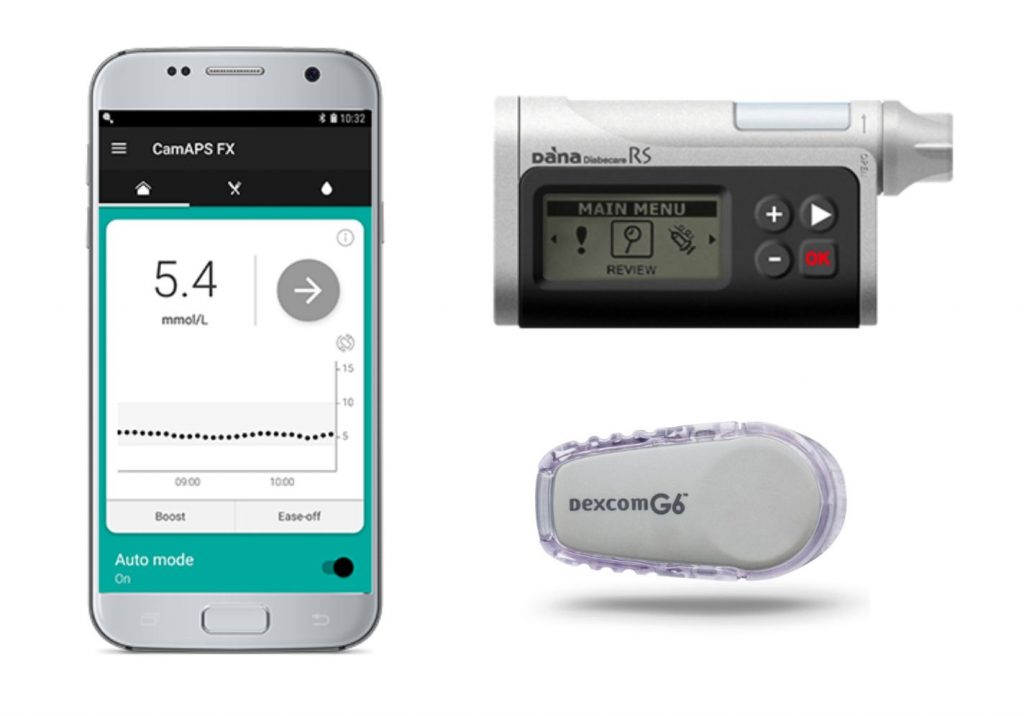
Writing in the New England Journal of Medicine, researchers compared the performance of the artificial pancreas, which uses an algorithm to determine the amount of insulin administered by a device worn by the child, against ‘sensor-augmented pump therapy’.
Management of type 1 diabetes is challenging in very young children, because of a number of factors including the high variability in levels of insulin required and in how individual children respond to treatment, and their unpredictable eating and activity patterns. Children are particularly at risk of dangerously low blood sugar levels (hypoglycaemia) and high blood sugar levels (hyperglycaemia). Previous studies have linked prolonged hyperglycaemia in children with type 1 diabetes with lower IQ scores and slower brain growth.
To manage children’s glucose levels, doctors increasingly turn to devices that continuously monitor glucose levels and deliver insulin via a pump, which administers insulin through a cannula inserted into the skin. These devices have proved successful to an extent in older children, but not in very young children.
Current technology – sensor-augmented pump therapy – requires parents to review their child’s glucose levels using a monitor and then manually adjust the amount of insulin administered by the pump.

Professor Roman Hovorka from the Wellcome-MRC Institute of Metabolic Science at the University of Cambridge, pictured left, has developed an app – CamAPS FX – which, combined with a glucose monitor and insulin pump, acts as an artificial pancreas, automatically adjusting the amount of insulin it delivers based on predicted or real-time glucose levels. It is a ‘hybrid closed loop system’, meaning that the child’s carer will have to administer insulin at mealtimes, but at all other times the algorithm works by itself. There are no commercially available versions of fully closed loop systems yet.
Professor Hovorka explained: “CamAPS FX makes predictions about what it thinks is likely to happen next based on past experience. It learns how much insulin the child needs per day and how this changes at different times of the day. It then uses this to adjust insulin levels to help achieve ideal blood sugar levels. Other than at mealtimes, it is fully automated, so parents do not need to continually monitor their child’s blood sugar levels.”
Working across seven centres in the UK and Europe, Professor Hovorka and an international team of researchers recruited 74 children with type 1 diabetes, aged one to seven years, to take part in their trial. The trial compared the safety and efficacy of hybrid closed-loop therapy with sensor-augmented pump therapy. All children used the CamAPS FX hybrid closed-loop system for 16 weeks, and then used the control treatment (sensor-augmented pump therapy) for 16 weeks.
On average, children spent around three-quarters of their day (71.6%) in the target range for their glucose levels when using CamAPS FX – almost nine percentage points higher compared to the control period, accounting for an additional 125 minutes per day in the target range.
The children spent less than a quarter (22.9%) of their time with raised blood sugar levels – hyperglycemia – while using CamAPS FX, almost nine percentage points lower than during the control period. There was no difference between the two groups in the time spent in hypoglycemia.
The app reduced average blood sugar levels – a measurement of a molecule known as glycated haemoglobin, or HbA1c. Glycated haemoglobin develops when haemoglobin, a protein within red blood cells that carries oxygen throughout the body, joins with glucose in the blood, becoming ‘glycated’. By measuring HbA1c, clinicians are able to get an overall picture of what a person’s average blood sugar levels have been over a period of weeks or months. For people with diabetes, the higher the HbA1c, the greater the risk of developing diabetes-related complications.
At baseline, average HbA1c levels were 7.3% – the app reduced this by 0.7 percentage points. This is particularly noteworthy as the study participants had good glycaemic control – that is, relatively low HbA1c – to begin with, and it is often hard to improve glucose control without having more low blood glucose events (hypoglycaemia).
Dr Julia Ware, the study’s first author, also from the Wellcome-MRC Institute of Metabolic Science, said: “Very young children are extremely vulnerable to changes in their blood sugar levels. High levels in particular can have potentially lasting consequences to their brain development. On top of that, diabetes is very challenging to manage in this age group, creating a huge burden for families.
“CamAPS FX led to improvements in several measures, including hyperglycaemia and average blood sugar levels, without increasing the risk of hypos. This is likely to have important benefits for those children who use it.”
One of the biggest challenges reported by families of young children with type 1 diabetes is poor sleep quality, as variability in insulin requirements and parental fear of hypoglycemia are highest overnight. In their study, the researchers found that more than 80% of overnight sensor readings were within the target range, showing that hybrid closed-loop therapy addresses the ‘night-time problem’ more effectively than sensor-augmented pump therapy.
Dr Ware added: “Parents have described our artificial pancreas as ‘life-changing’ as it meant they were able to relax and spend less time worrying about their child’s blood sugar levels, particularly at night time. They tell us it gives them more time to do what any ‘normal’ family can do, to play and do fun things with their children.”
CamAPS FX is already having an impact on the lives of children and their families. It is available through a number of NHS trusts across the UK, including Cambridge University Hospitals NHS Foundation Trust, and the team hope it will soon be available even more widely.
Professor Hovorka added: “From the first clinical trials of our algorithms to today’s findings has taken well over a decade, but the dedication of my team and the support of all the children and families who have taken part in our studies, has paid off. We believe our artificial pancreas will transform the lives of families with very young children affected by type 1 diabetes.”
CamAPS FX has been shown to work in older children and adolescents with type 1 diabetes. Today’s study is the first time that it has been shown to be effective over several months in very young children.
The research was funded by the European Commission’s Horizon 2020 Framework Programme, with additional support from the NIHR Cambridge Clinical Research Facility and JDRF.

“I feel like for the first time since the diagnosis I can relax”: Sofia and Sam’s story
Over the past three years Sam Wright, mother to Sofia (aged 6), pictured above, has endured the tremendously steep learning curve any parent of a child with type 1 diabetes has to undergo. A whirlwind of finger prick tests, injections and sensors later, she has now discovered the CamAPS FX app and the hybrid closed-loop system and would not be without it.
In a hot summer, excessive thirst wouldn’t be anything out of the ordinary for a young child. However, Sam and her mother followed their intuition that Sofia’s condition could be something more serious and began to research if there was any cause for concern, which ultimately led to her diagnosis of type 1 diabetes.
“The diagnosis changed everything forever,” says Sam. “I can remember the first few weeks like it was yesterday. Almost overnight I felt like I needed to be an expert in diabetes to best care for my little girl. She is my absolute priority, so I just did it, and I feel really proud of myself and my mum for doing it – she’s been on this journey with us from the beginning.”
A steep learning curve
Like most parents with a newly diagnosed child with type 1 diabetes, Sam quickly became an expert at finger prick tests, basal and bolus insulin dosing and what to do when her daughter was hypoglycaemic. Injecting your child would be a challenge for any parent, but Sam also had to overcome her own fear of needles.
“The clinical team at Addenbrooke’s Hospital were amazing at helping me overcome some big challenges in those early days, especially Adam Dawes, Clinical Nurse Specialist at the hospital. I don’t think we’d be where we are today without the support of Adam and the clinical team.”
In the first few months of Sofia’s diagnosis, finger prick tests were a regular occurrence to understand blood glucose levels throughout the day. Sam also had to set alarms at night so she could check her daughter’s blood glucose levels. If Sofia’s levels were high or low, Sam would have to administer a corrective dose and then wait until her levels were moving in the right direction before going back to sleep, an exhausting process for both every single night.
Eventually Sofia received a continuous glucose monitor (CGM) to support her care. This is a small device with a sensor worn just under the skin to measure glucose levels continuously. Sam saw the CGM as a step in the right direction, particularly as the device meant she no longer had to set alarms at night – the device would alert her automatically if Sofia’s glucose levels were out of range. It also syncs with Sam’s phone so she can check Sofia’s blood glucose levels at school and it also helps teachers with care through the day.
Finding freedom
In January 2020 Sam was introduced to the CamAPS FX app. She hasn’t looked back since.
Sam had never heard of the hybrid closed-loop system, or the CamAPS FX app before, but when Sofia was invited to take part in the KidsAP research trial, led by the University of Cambridge, she “jumped in with both feet”.
“I have full trust in the CamAPS FX app and I feel like for the first time since the diagnosis I can relax,” she says. The time Sofia spends within her target blood glucose range has improved and it is much easier now to control her levels. “It’s a complete weight off my shoulders.”
The app has also reduced the burden for Sofia’s teachers. Sam is able to check on her daughter remotely, but has the reassurance that she will receive automated text messages if Sofia’s glucose levels are going high or low.
“You wouldn’t know that she is any different from any of her classmates and that is thanks to the CamAPS FX app.”
Sam says it makes sense for children of Sofia’s age to have the closed-loop, because their bodies are constantly changing how they respond to insulin. It is an extremely difficult ask for a parent to manage their child’s condition without significant highs and lows, whereas the app learns and adapts instantly. It is beneficial for the child’s long-term management of the condition and enables both children and their parents to sleep at night.
“I would never be without the app,” says Sam. “It is something looking out for you so you don’t have to worry. To anyone considering it, just go for it. It is a game changer and you won’t look back or want to be without it once you’ve had it. I completely believe it is so beneficial for both the parents and the child.”
Adapted from University of Cambridge press release.
Photo of Sam Wright and Sofia, credit: Phil Mynott.
Award given to build data ‘bridge’ that could revolutionise precision medicine
NIHR Cambridge Biomedical Research Centre (NIHR Cambridge BRC), together with Genomics England (GEL), Eastern Academic Health Science Network (Eastern AHSN) and the precision medicine software company Lifebit have been awarded £200,000 by UK Research and Innovation as part of the DARE UK (Data and Analytics Research Environments UK) programme, to develop and test a ‘bridge’ between health data at the NIHR Cambridge BRC and GEL’s clinical genetic data – which will allow researchers to work with their combined data, without any data leaving either source.
“The more health data available for research, the more powerful it is – and through the NHS and research organisations such as NIHR and GEL, the UK has a lot of biomedical research data that could hold the key to understanding, diagnosing, and treating health conditions.

“However, data are often locked up in different locations due to the size of the data and to maintain privacy and security, ultimately preventing researchers from using it to its full potential,” explains Professor Serena Nik-Zainal, consortium lead and Genomic Medicine theme lead at NIHR Cambridge BRC, pictured right.
Organisations often store their data in spaces known as ‘Trusted Research Environments’ (TREs) – secure spaces for researchers to access and analyse sensitive data to help prevent unauthorised access and re-identification of individuals from anonymised data.
The DARE UK award will bring together a consortium to create and test a ‘bridge’ that will enable their respective TRE’s to ‘talk’ to one another (known as ‘federation’). Importantly, this bridging technology will be open source, meaning the global research community will be able to benefit from the collaborative potential of this technology.
Lifebit is already successfully working with GEL, having launched in 2020 a next-generation genomic medicine research platform that has been central to the UK Government’s research response to COVID-19, as well as facilitating medical advancements in cancer and rare diseases. This will be key to bridging the two datasets of Genomics England and the NIHR Cambridge BRC, in what will be the first federated architecture between a national project and a higher education institution.

As well as designing and testing the ‘bridging’ Federation infrastructure, the project aims to develop new standards to inform how federated TREs communicate securely and power largescale research analyses going forward.
All health data ultimately comes from patients themselves, and patient and public involvement has been essential from the outset. Consortium patient partner Rosanna Fennessy explains: “Patients also need answers about their conditions that hopefully patient data can provide. It is important to bring together all forms of expertise – patients, clinicians, researchers and data experts to design ways to safely and securely maximise the use of data for research.” The consortium will work with Rosanna, and patient groups across the UK to develop data governance and ethics frameworks, and federation best practices.
Professor Serena Nik-Zainal concludes: “We want to use health data to fix real, human problems and to narrow the gap between clinicians and computational experts working on health data for research.
“This starts with building the infrastructure, the machinery we need to be able to take advantage of the amazing data resources that we have to realise the potential of the data to improve patient lives.”
Find out more in this short video with Professor Serena Nik-Zainal.
Omicron may be significantly better at evading vaccine-induced immunity, but less likely to cause severe disease
The Omicron variant of SARS-CoV-2 may be significantly better than previous variants at evading vaccine-induced antibodies, according to new research from Cambridge – but preliminary evidence suggests it is less likely to cause severe COVID-19 illness in the lungs.
As the SARS-CoV-2 virus replicates and spreads, errors in its genetic code can lead to changes in the virus. On 26 November 2021, the World Health Organization designated the variant B.1.1.529, first identified in South Africa, a variant of concern, named Omicron. The variant carries a large number of mutations, leading to concern that it will leave vaccines less effective at protecting against infection and illness.
Working in secure conditions, a team led by Professor Ravi Gupta at the Cambridge Institute of Therapeutic Immunology and Infectious Disease, University of Cambridge, created synthetic viruses – known as ‘pseudoviruses’ – that carried key mutations found in the Delta and Omicron strains. They used these to study the virus’s behaviour.
The team, which included collaborators from Japan, including Dr Kei Sato of the University of Tokyo, has released its data ahead of peer review because of the urgent need to share information relating to the pandemic, and particularly the new Omicron variant.
Professor Gupta and colleagues tested the pseudoviruses against blood samples donated to the NIHR COVID-19 BioResource. The blood samples were from vaccinated individuals who had received two doses of either the AstraZeneca (ChAdOx-1) or Pfizer (BNT162b2) vaccines.
On average, Omicron required around a ten-fold increase in the concentration of serum antibody in order to neutralise the virus, compared to Delta. Of particular concern, antibodies from the majority of individuals who had received two doses of the AstraZeneca vaccine were unable to neutralise the virus. The data were confirmed in live virus experiments.
Reassuringly, however, following a third dose of the Pfizer vaccine, both groups saw a significant increase in neutralisation.
Professor Gupta said: “The Omicron variant appears to be much better than Delta at evading neutralising antibodies in individuals who have received just two doses of the vaccine. A third dose ‘booster’ with the Pfizer vaccine was able to overturn this in the short term, though we’d still expect a waning in immunity to occur over time.”
Spike proteins on the surface of SARS-CoV-2 bind to ACE2, a protein receptor found on the surface of cells in the lung. Both the spike protein and ACE2 are then cleaved, allowing genetic material from the virus to enter the host cell. The virus manipulates the host cell’s machinery to allow the virus to replicate and spread.
To see how effective Omicron is at entering our cells, the team used their pseudoviruses to infect cells in lung organoids – ‘mini-lungs’ that model parts of the lung. Despite having three mutations that were predicted to favour the spike cleavage, the researchers found the Omicron spike protein to be less efficient than the Delta spike at cleaving the ACE2 receptor and entering the lung cells.
In addition, once Omicron had entered the cells, it was also less able than Delta to cause fusion between cells, a phenomenon associated with impaired cell-to-cell spread. Fused cells are often seen in respiratory tissues taken following severe disease. Indeed, when the team used a live Omicron virus and compared it to Delta in a spreading infection experiment using lung cells, Omicron was significantly poorer in replication, confirming the findings regarding impaired entry.
Professor Gupta added: “We speculate that the more efficient the virus is at infecting our cells, the more severe the disease might be. The fact that Omicron is not so good at entering lung cells and that it causes fewer fused cells with lower infection levels in the lab suggests this new variant may cause less severe lung-associated disease.
“While further work is needed to corroborate these findings, overall, it suggests that Omicron’s mutations present the virus with a double-edged sword: it’s got better at evading the immune system, but it might have lost some of its ability to cause severe disease.”
However, Professor Gupta urged caution.
“Omicron still represents a major public health challenge. Individuals who have only received two doses of the vaccine – or worse, none at all – are still at significant risk of COVID-19, and some will develop severe disease. The sheer number of new cases we are seeing every day reinforces the need for everyone to get their boosters as quickly as possible.”
The research was supported by Wellcome and the NIHR Cambridge Biomedical Research Centre.
Paper Reference
Meng, B, et al. SARS-CoV-2 Omicron neutralising antibody evasion, replication and cell-cell fusion.
NICE prostate guidelines updated based on Cambridge research
A Cambridge developed model to risk-categorise prostate cancer has been added to the NICE guidelines and adopted as standard practice in the NHS.
Patients who are newly diagnosed with prostate cancer are currently filtered into three groups: low, intermediate or high-risk. A new risk model developed in Cambridge – the Cambridge Prognostic Groups (CPG), has shown re-categorising these groups into five categories, based on more detailed information is better able to predict disease aggressiveness and benefit patient care.
Developed by the University of Cambridge, Cambridge University Hospitals and supported by the NIHR Cambridge BRC and CRF, researchers have demonstrated that having clearer, detailed groups will help monitor and manage the disease.
The National Institute for Health and Care Excellence (NICE), who provides national guidance and advice for health, public health and social care practitioners, have recognised this new grouping system as an improved method when deciding on treatments for newly diagnosed patients and have now amended their guidelines for treating prostate cancer.
Re-evaluating risk categories for prostate cancer
The prostate is a gland located underneath the bladder and surrounds the urethra (tube that carries urine). This gland can grow and could potentially turn cancerous. The causes of prostate cancer are largely unknown and signs of the disease include blood in urine or semen, difficulty urinating or bone pain.
Those referred with a raised blood level of Prostate Specific Antigen (PSA) or other worrying symptoms have tests to determine any presence of cancer. If they are diagnosed with the disease, they were normally categorised in one of three groups, depending on the severity of the cancer, and start treatment.

However, new research from Professor Vincent J Gnanapragasam, Professor of Urology, pictured right, and the team demonstrated that some patients did not fit neatly into any of the categories and so did not require the same monitoring and treatments compared to others in the respective groups.
After years of collecting more detailed health information and testing, the team found having five categories – Cambridge Prognostics Groups 1-5– was a better method to inform the best treatment choice and also likelihood of the disease responding to treatment.
From this extensive research, NICE have now recognised all newly diagnosed men in the UK should be risk-categorised by the new five category model developed in the CPG’s criteria. This will lead to better care and tailoring appropriate treatment to patients.
Professor Gnanapragasam said: “The current three-tier model has been used for over twenty years, we’ve come a long way since then and have a better understanding of prostate cancer. Having this new model will make sure we are able to better predict how men with similar risk types will respond to treatment. More importantly it will also reduce the amount of unnecessary treatments for those where the disease is unlikely to cause problems during a man’s natural lifespan.
“Having our model independently reviewed and adopted by NICE and now recommended in mainstream treatment is a testament to the robustness of our research all the hard work we have done to improve how we manage prostate cancer.”
Com-COV 3 COVID-19 vaccine study calls on teenager volunteers in Cambridgeshire
Researchers running the University of Oxford-led Com-COV programme have started enrolling young people aged 12 to 16 years old to receive a second dose of a COVID-19 vaccine, including in Cambridgeshire.
Backed through funding from the Vaccines Taskforce and National Institute for Health Research (NIHR) and run across several NIHR-supported sites by the National Immunisation Schedule Evaluation Consortium, the Com-COV 3 trial is seeking to recruit 270 volunteers. The study has already opened at six sites in the UK, and is expanding to nine new recruiting areas, with recruitment happening locally at NIHR Cambridge Clinical Research Facility at Cambridge University Hospitals NHS Foundation Trust (CUH).
Professor Matthew Snape, Associate Professor in Paediatrics and Vaccinology at the Oxford Vaccine Group, and Chief Investigator on the trial, said: “Teenagers are currently experiencing the highest rate of infections of all age groups in the UK. This study will be critical to delivering vital information on the range of options for immunising teenagers against COVID-19 in the UK to help control this. Therefore, we are asking for 12 to 16 years old to take this opportunity to receive a second dose of vaccine and help us understand how best to immunise teenagers to protect them and their families.”
All participants will be randomly allocated at the time of their second dose to receive either a full second dose of the Pfizer vaccine, a one-third dose of the Pfizer vaccine, or a full dose of the Novavax vaccine. These vaccines will be administered at least eight weeks after their first dose.
The current UK guidance is that all 12 to 15 year olds receive a single dose of vaccine, while 16 to 17 year olds receive 2 doses of vaccine, 12 weeks apart. Younger people at a greater risk of serious illness if they catch COVID-19 are currently offered two doses. The results from the study will provide the JCVI (Joint Committee on Vaccination and Immunisation) with timely and crucial information about immunising teenagers in the UK.
Dr Theofilos Polychronakis, consultant in paediatric respiratory medicine, who is leading the trial at CUH, said: “Participation in research by people of Cambridgeshire has always contributed enormously to the discovery of new treatments, particularly over the last two years, and we are extremely grateful to them and all those who help our researchers to make a difference.
“It’s very important that a broad range of people from all parts of the country, and of all ages, take part in research in order to find the most effective solutions to COVID-19 possible. Therefore, we welcome 12-16 year olds from our local community to take part in this study and help us determine the best options for immunisation for all teenagers, everywhere.”
Professor Matthew Snape said: “We are very grateful to those young volunteers and their parents who have signed up for the study so far. We hope the addition of the trial site at CUH will encourage even more participants to get involved in this critically important research.”
The study is single-blind and randomised, meaning participants will not know what second dose vaccine they are receiving. Researchers will assess reactogenicity (any side effects) and immune system responses to these new combinations of vaccines.
Professor Andrew Ustianowski, NIHR Clinical Lead for COVID-19 Vaccination Programme and Joint National Infection Specialty Lead, said: “By getting involved in this study, volunteers will be able to help researchers develop our understanding of how we can best protect teenagers against COVID-19.
“Thanks to the generosity of thousands of vaccine study participants over the past 18 months, we have been able to reduce the impact and spread of COVID-19 with approved vaccines. Once it has reached its target, Com-COV 3 will be a pivotal study that is expected to provide important data that will lead directly to UK guidance on protecting young people and their families.”
If you would like to register to take part in the study at CUH, visit the study website.
Using air filters on hospital wards remove almost all airborne Covid virus
A new study has found placing air filtration machines in COVID-19 wards at Addenbrooke’s Hospital, removed almost all traces of airborne SARS-CoV-2 virus.
Supported by the NIHR Cambridge BRC, the research was led by doctors, scientists and engineers at Addenbrooke’s and the University of Cambridge in January, at the height of the second wave of the pandemic.
This discovery could have implications for improving the safety of repurposed ‘surge wards’, the researchers say it also opens up the possibility of being able to set standards for cleaner air to reduce the risk of airborne transmission of infections.
Over the duration of the pandemic there has been a steady rise in the evidence that the SARS-CoV-2 virus can be transmitted through the air in small droplets (aerosols). But as hospitals have seen their capacity overwhelmed, they have been forced to manage many of their COVID-19 patients in repurposed ‘surge’ wards, which often lack the ability to change the air with a high frequency. While the use of appropriate personal protective equipment (PPE) protects staff and patients significantly reduces the risk of transmission, there are still reports of patient-to-healthcare worker transmission of the virus, potentially through the inhalation of viral particles.
A team at the University of Cambridge and Cambridge University Hospitals (CUH) NHS Foundation Trust investigated whether portable air filtration/UV sterilisation devices could reduce airborne SARS- CoV-2 in general wards that had been repurposed as a COVID ward and a COVID Intensive Care Unit (ICU). The results are published in Clinical Infectious Diseases.
Dr Vilas Navapurkar, a Consultant in Intensive Care Medicine at CUH, who led the study, said: “Reducing airborne transmission of the coronavirus is extremely important for the safety of both patients and staff. Effective PPE has made a huge difference, but anything we can do to reduce the risk further is important.”
“Because of the numbers of patients being admitted with COVID-19, hospitals have had to use wards not designed for managing respiratory infections. During an intensely busy time, we were able to pull together a team from across the hospital and University to test whether portable air filtration devices, which are relatively inexpensive, might remove airborne SARS-CoV-2 and make these wards safer.”
The team performed their study in two repurposed COVID-19 units in Addenbrooke’s Hospital. One area was a surge ward managing patients who required simple oxygen treatment or no respiratory support; the second was a surge ICU managing patients who required ventilation either through non-invasive mask ventilation or invasive respiratory support, such as involving the use of an invasive tube and tracheostomy.
The team installed a High Efficiency Particulate Air (HEPA) air filter/UV steriliser. HEPA filters are made up of thousands of fibres knitted together to form a material that filters out particles above a certain size. The machines were placed in fixed positions and operated continuously for seven days, filtering the full volume of air in each room between five and ten times per hour.
In the surge ward, during the first week prior to the air filter being activated, the researchers were able to detect SARS-CoV-2 on all sampling days. Once the air filter was switched on and run continuously, the team were unable to detect SARS-CoV-2 on any of the five testing days. They then switched off the machine and repeated the sampling – once again, they were able to detect SARS-CoV-2 on three of the five sampling days.
On the ICU, the team found limited evidence of airborne SARS-CoV-2 in the weeks when the machine was switched off and traces of the virus on one sampling day when the machine was active.
Additionally, the air filters significantly reduced levels of bacterial, fungal and other viral bioaerosols on the both the surge ward and the ICU, highlighting an added benefit of the system.
First author Dr Andrew Conway Morris, from the Department of Medicine at the University of Cambridge, said: “We were really surprised by quite how effect air filters were at removing airborne SARS-CoV-2 on the wards. Although it was only a small study, it highlights their potential to improve the safety of wards, particularly in areas not designed for managing highly infectious diseases such as COVID-19.”
Crucially, the research team developed a robust technique for assessing the quality of air, involving placing air samplers at various points in the room and then testing the samples using PCR assays similar those used in the ‘gold standard’ COVID-19 tests.
Professor Stephen Baker, from the Cambridge Institute of Therapeutic Immunology and Infectious Disease at the University of Cambridge, said: “Cleaner air will reduce the risk of airborne disease transmission, but it’s unlikely to be the case that just installing an air filter will be enough to guarantee the air is clean enough. Every room and every situation will be different. A key part of our work has been developing a robust way of measuring air quality.”
Dr Navapurkar added: “We’re all familiar with the idea of having standards for clean water and of hygiene standards for food. We need now to agree standards for what is acceptable air quality and how we meet and monitor those standards.”
The research was also supported by Wellcome and the Medical Research Council.
Adapted from University of Cambridge release
New ground-breaking research that is a potential game-changer for diagnosis and treatment of undiagnosed rare disease patients
In a world-first, new research published in the New England Journal of Medicine, has shown that whole genome sequencing (WGS) can uncover new diagnoses for patients with rare diseases – with potential to deliver enormous benefits across the NHS.
Led by Genomics England and Queen Mary University of London, researchers in this pilot study of rare undiagnosed diseases invited participants and their families from the 100,000 Genomes Project and NIHR BioResource to provide blood samples for WGS (whole genome sequencing), which reads a person’s entire genetic code.
After analysing the genes of 4,660 people with a rare disease from 2,183 families using WGS, researchers were able to find a genetic diagnosis for a quarter of them. Researchers also found that 14% of people who received a diagnosis had their disease identified in regions of their genome that other standard tests may have missed.
Uncovering a diagnosis
According to Rare Disease UK, 1 in 17 people will be affected by a rare disease – that’s 3.5 million people in the UK and 30 million people across Europe.
But it can take years of rigorous tests and appointments for patients to find out what their condition is – and sometimes they may never know.
By having their whole genome sequenced, diagnoses were uncovered that previous tests could not detect. For 25% of study participants, their diagnosis means they can now receive more focused clinical care – including further family screening, dietary change, vitamin / mineral provision and other therapies.
This ground-breaking research doesanalyse the diagnostic and clinical impact of WGS for a broad range of rare diseases within a national healthcare system – and has shown how it can effectively secure a diagnosis for patients and save the NHS vital resources.
Participants who received a diagnosis through the pilot include:
- A 10-year-old girl whose previous seven-year search for a diagnosis had multiple intensive care admissions over 307 hospital visits at a cost of £356,571. Genomic diagnosis enabled her to receive a curative bone marrow transplant (at a cost of £70,000). In addition, predictive testing of her siblings showed no further family members were at risk.
- A man in his 60s who had endured years of treatment for a serious kidney disease, including two kidney transplants. Already knowing his daughter had inherited the same condition, a genomic diagnosis made by looking at the whole genome for him and his daughter enabled his 15-year-old granddaughter to be tested. This revealed she had not inherited the disease and could cease regular costly check-ups.
- A baby who became severely ill immediately after birth and sadly died at four months but with no diagnosis and healthcare costs of £80,000. Analysis of the baby’swhole genome uncovered a severe metabolic disorder due to inability to take vitamin B12 inside cells explaining his illness. This enabled a predictive test to be offered to the younger brother within one week of his birth. The younger child was diagnosed with the same disorder but was able to be treated with weekly vitamin B12 injections to prevent progression of the illness.
Professor Sir Mark Caulfield (lead author) from Queen Mary University of London, and former Chief Scientist at Genomics England, said: “We hope this major advance will enable rare disease patients worldwide to start receiving diagnostic whole genome sequencing where appropriate. Our findings show that deployment of this comprehensive and efficient genomic test at the first signs of symptoms, can improve diagnostic rates. This study has paved the way for clinical implementation of whole genome sequencing as part of the NHS Genomic Medicine Service.”
Professor Damian Smedley (lead author) from Queen Mary University of London, explained: “This is the first time that whole genome sequencing has been directly embedded into rare disease diagnostics in a healthcare system like the NHS and applied at scale across the full breadth of rare disease. Our novel software, together with collection of detailed clinical data, was key to us being able to solve the “needle in a haystack” challenge of finding the cause of a rare disease patient’s condition amongst the millions of variants in every genome. A large proportion of the diagnoses we discovered were found outside the coding region and would not have been detected by existing approaches. This study makes the case for healthcare systems worldwide to adopt whole genome sequencing as the genetic test of choice for rare disease patients.”
Professor John Bradley, Chief Investigator of the NIHR BioResource, added: “The NIHR BioResource is delighted to have worked in partnership with Genomics England and NHS England to deliver this study. It is transforming the approach to diagnosing rare diseases in the NHS.”
The study was also conducted in partnership with the National Institute for Health Research (NIHR) and Illumina who undertook the sequencing, and it was funded by the NIHR, the Wellcome Trust, the Medical Research Council, Cancer Research UK, the Department of Health and Social Care, and NHS England.
NIHR Biomedical Research Centres at Barts, Cambridge, Great Ormond Street Hospital for Children NHS, Manchester, Moorfields, Newcastle, Oxford and University College London Hospitals supported this research
